Identification and Evolution of LRR VIII-2 Subfamily Genes in Four Model Plant Species
- 1Key Laboratory of Forest Genetics and Biotechnology of Ministry of Education, Nanjing Forestry University, Nanjing 210037, China
2Co-Innovation Center for the Sustainable Forestry in Southern China, College of Forestry, Nanjing Forestry University, Nanjing 210037, China
Received date: 2019-08-16
Accepted date: 2020-03-23
Online published: 2020-03-23
Abstract
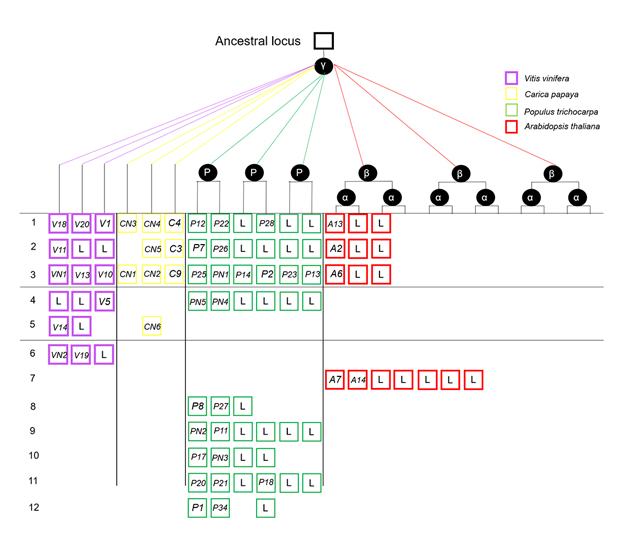
Whole-genome duplication and tandem duplication are two important mechanisms for gene duplication, which play important roles in promoting the genomic and genetic diversity. In Arabidopsis, AtLRR-RLK encodes receptor-like kinases rich in leucine repeats, which is a multi-gene family arising from large-scale gene expansion during angiosperm evolution. It is composed of 15 subfamilies, among which, AtLRR VIII-2 is the subfamily with the highest proportion of tandem repeats. In this study, we use the genes in LRR VIII-2 as an example to analyze the gene expansion and differential retention in four model plants (Arabidopsis thaliana, Populus trichocarpa, Vitis vinifera and Carica papaya). Results showed that paralogous gene pairs were identified in the LRR VIII-2 subfamily in Arabidopsis, poplar and grape, while no such pair was found in papaya. The LRR VIII-2 subfamily expanded the most significantly in poplar and moderately expanded in Arabidopsis and grape, but some genes of the LRR VIII-2 subfamily in papaya have been lost. In addition, the paralogous and orthologous genes in the LRR VIII-2 subfamily were under strong purifying selection in the four investigated plant species, except for a pair of paralogous genes in poplar. An in-depth phylogenetic analysis of the LRR VIII-2 subfamily helps to understand the role and significance of gene duplication in plant evolution, and provides useful information for predicting the function of homologous gene among different species. This analytical pipeline is also applicable for deciphering the evolution history of other gene families.
Cite this article
Chenyang Yan , Yingnan Chen . Identification and Evolution of LRR VIII-2 Subfamily Genes in Four Model Plant Species[J]. Chinese Bulletin of Botany, 2020 , 55(4) : 442 -456 . DOI: 10.11983/CBB19157
References
| [1] | 孙红正, 葛颂 (2010). 重复基因的进化——回顾与进展. 植物学报 45, 13-22. |
| [2] | 王鹤飞, 李雪, 董玲丽, 张俊成, 赵茂林, 邢国珍, 王道文, 郑文明 (2015). 小麦受体样蛋白激酶及其衍生蛋白的研究进展. 植物学报 50, 255-262. |
| [3] | 闫锋, 祝传书, 庞保平 (2009). 植物类受体蛋白激酶的研究进展. 西北植物学报 29, 851-858. |
| [4] | 张兆沛, 王志伟, 张慧蓉 (2009). 植物类受体蛋白激酶研究概况. 山西农业科学 37, 75-78. |
| [5] | 郑超, 李登高, 白薇 (2016). 植物富含半胱氨酸的类受体激酶的研究进展. 生物技术通报 32, 10-17. |
| [6] | Albrecht C, Russinova E, Hecht V, Baaijens E, de Vries S (2005). The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES 1 and 2 control male sporogenesis. Plant Cell 17, 3337-3349. |
| [7] | Bej A, Sahoo BR, Swain B, Basu M, Jayasankar P, Samanta M (2014). LRRsearch: an asynchronous server- based application for the prediction of leucine-rich repeat motifs and an integrative database of NOD-like receptors. Comput Biol Med 53, 164-170. |
| [8] | Blanc G, Hokamp K, Wolfe KH (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res 13, 137-144. |
| [9] | Blanc G, Wolfe KH (2004). Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16, 1667-1678. |
| [10] | Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2002). TBtools, an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13, 1194-1202. |
| [11] | Clark SE, Williams RW, Meyerowitz EM (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575-585. |
| [12] | Colcombet J, Boisson-Dernier A, Ros-Palau R, Vera CE, Schroeder JI (2005). Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES 1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 17, 3350-3361. |
| [13] | Conant GC, Wolfe KH (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9, 938-950. |
| [14] | Cummings MP (2004). PAUP* (phylogenetic analysis using parsimony (and other methods)). Santa Monica: American Cancer Society. pp. 37-45. |
| [15] | Dai XG, Hu QJ, Cai QL, Feng K, Ye N, Tuskan GA, Milne R, Chen YN, Wan ZB, Wang ZF, Luo WC, Wang K, Wan DS, Wang MX, Wang J, Liu JQ, Yin TM (2014). The willow genome and divergent evolution from poplar after the common genome duplication. Cell Res 24, 1274-1277. |
| [16] | Feng C, Mackey AJ, Vermunt JK, Roos DS (2007). Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS One 2, e383. |
| [17] | Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M (2014). Pfam: the protein families database. Nucleic Acids Res 42, D222-D230. |
| [18] | Finn RD, Clements J, Eddy SR (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39, W29-W37. |
| [19] | Fischer I, Diévart A, Droc G, Dufayard JF, Chantret N (2016). Evolutionary dynamics of the leucine-rich repeat receptor-like kinase (LRR-RLK) subfamily in angiosperms. Plant Physiol 170, 1595-1610. |
| [20] | Freeling M (2009). Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol 60, 433-453. |
| [21] | Gabaldón T, Dessimoz C, Huxley-Jones J, Vilella AJ, Sonnhammer EL, Lewis S (2009). Joining forces in the quest for orthologs. Genome Biol 10, 403. |
| [22] | Gómez-Gómez GL, Boller T (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5, 1003-1011. |
| [23] | Guo LH, Chen YN, Ye N, Dai XG, Yang WX, Yin TM (2014). Differential retention and expansion of the ancestral genes associated with the paleopolyploidies in modern rosid plants, as revealed by analysis of the extensins super-gene family. BMC Genomics 15, 612. |
| [24] | Hwang SG, Kim DS, Jang CS (2011). Comparative analysis of evolutionary dynamics of genes encoding leucine- rich repeat receptor-like kinase between rice and Arabidopsis. Genetics 139, 1023. |
| [25] | Jia YX, Ding YL, Shi YT, Zhang XY, Gong ZZ, Yang SH (2016). The cbfs triple mutants reveal the essential functions of cbfs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol 212, 345-353. |
| [26] | Lee TH, Tang HB, Wang XY, Paterson AH (2012). PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res 41, D1152-D1158. |
| [27] | Lehti-Shiu MD, Shiu SH (2012). Diversity, classification and function of the plant protein kinase superfamily. Philos Trans R Soc B Biol Sci 367, 2619-2639. |
| [28] | Letunic I, Bork P (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46, D493-D496. |
| [29] | Li WH, Yang J, Gu X (2005). Expression divergence between duplicate genes. Trends Genet 21, 602-607. |
| [30] | Librado P, Rozas J (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451-1452. |
| [31] | Liu ZY, Jia YX, Ding YL, Shi YT, Li Z, Guo Y, Gong ZZ, Yang SH (2017). Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol Cell 66, 117-128. |
| [32] | Marchler-Bauer A, Lu SN, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke ZX, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang DC, Zhang NG, Zheng CJ, Bryant SH (2011). CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 39, D225-D229. |
| [33] | Moore RC, Purugganan MD (2003). The early stages of duplicate gene evolution. Proc Natl Acad Sci USA 100, 15682-15687. |
| [34] | Ogawa M, Shinohara H, Sakagami Y, Matsubayashi Y (2008). Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319, 294. |
| [35] | Rameneni JJ, Lee Y, Dhandapani V, Yu XN, Choi SR, Oh MH, Lim YP (2015). Genomic and post-translational modification analysis of leucine-rich-repeat receptor-like kinases in Brassica rapa. PLoS One 10, e0142255. |
| [36] | Shiu HS, Bleecker AB (2003). Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol 132, 530-543. |
| [37] | Shiu SH, Bleecker AB (2001a). Plant receptor-like kinase gene family: diversity, function, and signaling. Sci STKE 2001, re22. |
| [38] | Shiu SH, Bleecker AB (2001 b). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98, 10763-10768. |
| [39] | Shumayla, Sharma S, Kumar R, Mendu V, Singh K, Upadhyay SK (2016). Genomic dissection and expression profiling revealed functional divergence in Triticum aestivum leucine rich repeat receptor like kinases (TaLRRKs). Front Plant Sci 7, 1374. |
| [40] | Song DH, Li GJ, Song FM, Zheng Z (2008). Molecular characterization and expression analysis of OsBISERK1, a gene encoding a leucine-rich repeat receptor-like kinase, during disease resistance responses in rice. Mol Biol Rep 35, 275-283. |
| [41] | Sonnhammer ELL, Koonin EV (2002). Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet 18, 619-620. |
| [42] | Stone JM, Walker JC (1995). Plant protein kinase families and signal transduction. Plant Physiol 108, 451-457. |
| [43] | Sun XL, Wang GL (2011). Genome-wide identification, characterization and phylogenetic analysis of the rice LRR-kinases. PLoS One 6, e16079. |
| [44] | Tang HB, Bowers JE, Wang XY, Ming R, Alam M, Paterson AH (2008a). Synteny and collinearity in plant genomes. Science 320, 486-488. |
| [45] | Tang HB, Wang XY, Bowers JE, Ming R, Alam M, Paterson AH (2008b). Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res 18, 1944-1954. |
| [46] | Tang P, Zhang Y, Sun XQ, Tian DC, Yang SH, Ding J (2010). Disease resistance signature of the leucine-rich repeat receptor-like kinase genes in four plant species. Plant Sci 179, 399-406. |
| [47] | The Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796-815. |
| [48] | Tian WD, Skolnick J (2003). How well is enzyme function conserved as a function of pairwise sequence identity? J Mol Biol 333, 863-882. |
| [49] | Wei ZR, Wang JH, Yang SH, Song YJ (2015). Identification and expression analysis of the LRR-RLK gene family in tomato(Solanum lycopersicum) Heinz 1706. Genome 58, 121-134. |
| [50] | Yang SH, Zhang XH, Yue JX, Tian DC, Chen JQ (2008). Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Genet Genomic 280, 187-198. |
| [51] | Yu Y, Fuscoe JC, Zhao C, Guo C, Jia MW, Tao Q, Bannon DI, Lancashire L, Bao WJ, Du TT, Luo H, Su ZQ, Jones WD, Moland CL, Branham WS, Qian F, Ning BT, Li Y, Hong HX, Guo L, Mei N, Shi TL, Wang KY, Wolfinger RD, Nikolsky Y, Walker SJ, Duerksen- Hughes P, Mason CE, Tong WD, Thierry-Mieg J, Thierry-Mieg D, Shi LM, Wang C (2014). A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat Commun 5, 3230. |
| [52] | Zan YJ, Ji Y, Zhang Y, Yang SH, Song YJ, Wang JH (2013). Genome-wide identification, characterization and expression analysis of Populus leucine-rich repeat receptor-like protein kinase genes. BMC Genomics 14, 318. |