In Vitro Ubiquitination Assay for Plant Proteins
- 1 College of Life Sciences, Liaocheng University, Liaocheng 252000, China
2 College of Life Sciences, Shandong University, Qingdao 266237, China
3 Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China
4 University of Chinese Academy of Sciences, Beijing 100049, China
Received date: 2019-08-12
Accepted date: 2019-10-31
Online published: 2019-10-31
Abstract
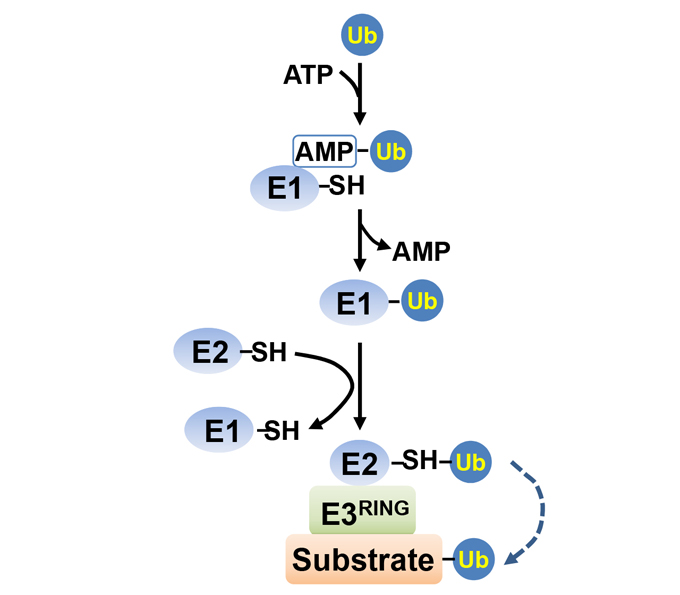
Ubiquitin activating enzyme (E1), ubiquitin conjugating enzyme (E2) and ubiquitin protein ligase (E3) are the key enzymes of ubiquitin modification of substrate proteins. There are large amounts of genes encoding these ubiquitination enzymes in all eukaryotic genomes. Analyzing the biochemical characteristics and specificity of these enzymes and their substrate proteins is important for their functional study. Here we describe a simple and fast method for in vitro ubiquitination assay. In the presence of E1 and ubiquitin, E2 activity can be determined by detecting the DTT-sensitive thio-ester formation. The E3 activity of a putative protein as well as the E2-E3 or E3-substrate specificities can also be explored by in vitro ubiquitination assay. This system is mainiy based on proteins from Arabidopsis, which includes most varieties of Arabidopsis E2 proteins that are tested with several RING-finger type E3 ligases. This system facilitate not only the exploration of E3 activity in combination with various Arabidopsis E2 members but also the study of E2-RING E3 and RING E3-substrate specificities. This system is suitable for the ubiquitination assays of eukaryotic proteins, especially for plant proteins.
Cite this article
Qingzhen Zhao , Lijing Liu , Qi Xie , Feifei Yu . In Vitro Ubiquitination Assay for Plant Proteins[J]. Chinese Bulletin of Botany, 2019 , 54(6) : 764 -772 . DOI: 10.11983/CBB19152
References
| [1] | 张祥云, 赵思语, 温潇, 王宁, 郭彦, 赵庆臻 ( 2018). 小麦TaUBC基因泛素结合酶活性分析. 聊城大学学报(自然科学版) 31, 79-85. |
| [2] | Bachmair A, Novatchkova M, Potuschak T, Eisenhaber F ( 2001). Ubiquitylation in plants: a post-genomic look at a post-translational modification. Trends Plant Sci 6, 463-470. |
| [3] | Callis J ( 2014). The ubiquitination machinery of the ubiquitin system. Arabidopsis Book 12, e0174. |
| [4] | Hua ZH, Vierstra RD ( 2011). The cullin-RING ubipuitinprotein ligases. Annu Rev Plant Biol 62, 299-334. |
| [5] | Kraft E, Stone SL, Ma LG, Su N, Gao Y, Lau OS, Deng XW, Callis J ( 2005). Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol 139, 1597-1611. |
| [6] | Michelle C, Vourc'h P, Mignon L, Andres CR ( 2009). What was the set of ubiquitin and ubiquitin-like conjugating enzymes in the eukaryote common ancestor? J Mol Evol 68, 616-628. |
| [7] | Miricescu A, Goslin K, Graciet E ( 2018). Ubiquitylation in plants: signaling hub for the integration of environmental signals. J Exp Bot 69, 4511-4527. |
| [8] | Proietto M, Bianchi MM, Ballario P, Brenna A ( 2015). Epigenetic and posttranslational modifications in light signal transduction and the circadian clock in Neurospora crassa. Int J Mol Sci 16, 15347-15383. |
| [9] | Smalle J, Vierstra RD ( 2004). The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55, 555-590. |
| [10] | Turek I, Tischer N, Lassig R, Trujillo M ( 2018). Multi-tiered pairing selectivity between E2 ubiquitin-conjugating enzymes and E3 ligases. J Bio Chem 293, 16324-16336. |
| [11] | Vierstra RD ( 2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci 8, 135-142. |
| [12] | Vierstra RD ( 2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10, 385-397. |
| [13] | Vierstra RD ( 2012). The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol 160, 2-14. |
| [14] | Xie Q, Guo HS, Dallman G, Fang SY, Weissman AM, Chua NH ( 2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419, 167-170. |
| [15] | Yu JW, Rubio V, Lee NY, Bai SL, Lee SY, Kim SS, Liu LJ, Zhang YY, Irigoyen ML, Sullivan JA, Zhang Y, Lee I, Xie Q, Paek NC, Deng XW ( 2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell 32, 617-630. |
| [16] | Zhang HW, Cui F, Wu YR, Lou LJ, Liu LL, Tian MM, Ning YS, Shu K, Tang SY, Xie Q ( 2015). The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell 27, 214-227. |
| [17] | Zhang YY, Yang CW, Li Y, Zheng NY, Chen H, Zhao QZ, Gao T, Guo HS, Xie Q ( 2007). SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19, 1912-1929. |
| [18] | Zhao QZ, Tian MM, Li QL, Cui F, Liu LJ, Yin BJ, Xie Q ( 2013). A plant-specific in vitro ubiquitination analysis system. Plant J 74, 524-533. |