核糖体图谱技术在植物学研究中的应用
- 中国科学院植物研究所, 中国科学院北方资源植物重点实验室, 北京 100093
收稿日期: 2022-02-14
录用日期: 2022-06-28
网络出版日期: 2022-06-28
基金资助
北京市自然科学基金(6212024);国家自然科学基金(32172638)
Recent Advances of Ribosome Profiling in Plants
- Key Laboratory of Plant Resources, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China
Received date: 2022-02-14
Accepted date: 2022-06-28
Online published: 2022-06-28
摘要
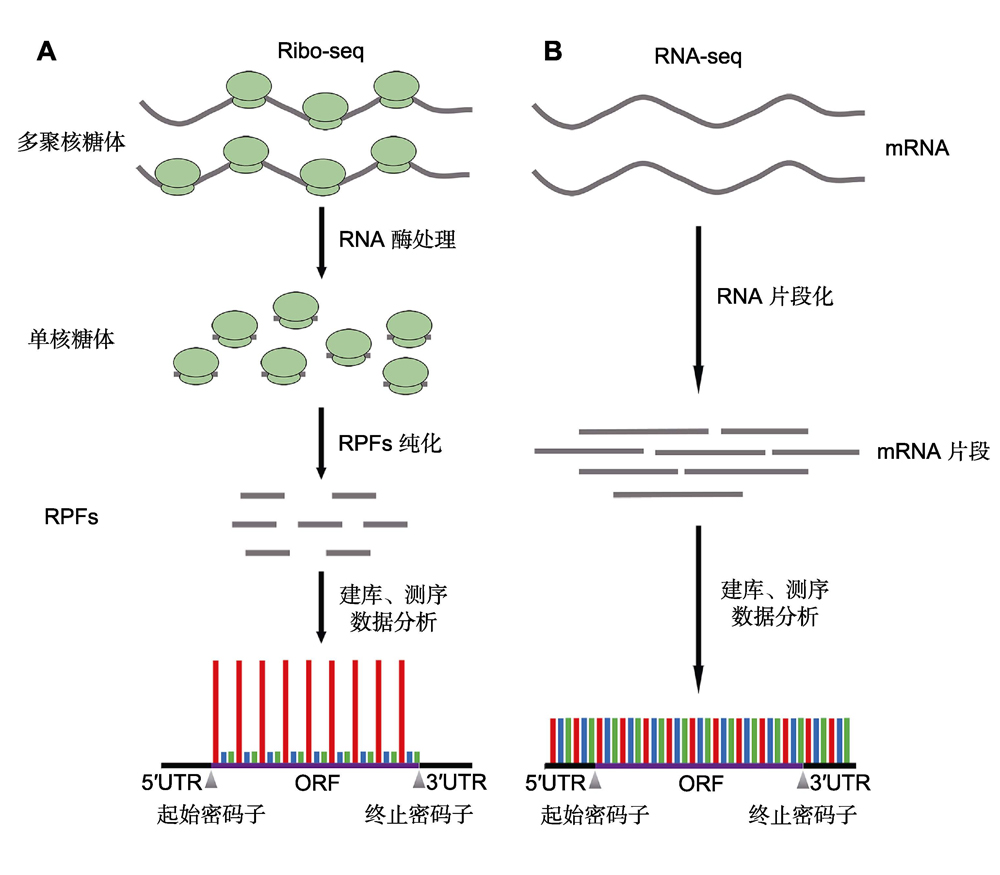
随着高通量测序技术的持续发展和进步, 开发出很多新颖的测序技术, 为诸多悬而未决的生物学难题提供了解决方案。其中, 核糖体图谱技术能够在全基因组水平和单核苷酸分辨率上监控细胞内的翻译事件, 填补了转录组学和蛋白质组学研究之间的空隙。核糖体图谱技术不仅能够鉴定处于翻译状态的RNA分子, 还能够精确定位RNA分子上正在翻译的核苷酸, 进而准确描绘RNA分子上的开放阅读框。此外, 结合转录组测序数据, 核糖体图谱技术还可以确定每个转录本上的核糖体数量, 从而计算每个转录本的翻译效率。目前, 核糖体图谱技术已成功应用于动物、植物和微生物等研究领域, 加深了人们对翻译调控机制的认识。然而, 由于植物细胞和组织的特性, 核糖体图谱技术在植物学研究中的应用仍然存在局限。该文综述了核糖体图谱技术的实验原理, 以及在植物学研究中的相关进展。
本文引用格式
王豫颖 , 王威浩 . 核糖体图谱技术在植物学研究中的应用[J]. 植物学报, 2022 , 57(5) : 673 -683 . DOI: 10.11983/CBB22028
Abstract
With the great development and progress of high-throughput sequencing (HTS) technologies, many newborn techniques have provided solutions to unresolved biological puzzles. Ribosome profiling has emerged as an HTS-based technique that facilitates the genome-wide investigation of translation at single-nucleotide resolution, and it resolves the missing link between transcriptome and translatome. Ribosome profiling not only identifies the mRNAs in translation, but also provides precise positions of the ribosomes on mRNA to help identify the coding regions of the mRNAs. In addition, combined with the parallel RNA sequencing, ribosome profiling can determine the translation efficiency for each mRNA by comparing the rates of protein synthesis and the abundance of mRNAs. Based on its extraordinary advances, ribosome profiling has been successfully applied in animals, plants and microbes, and has thoroughly enhanced our understanding of translational regulation. However, due to the restrictions caused by the physical characteristics of plant cells and tissues, there are still considerable limitations in the application of ribosome profiling in plants. Here, we introduce the experimental principle of ribosome profiling and review the recent progresses made in botanical researches.

参考文献
| [1] | 林忠平 (1984). 植物mRNA和多聚核糖体的分离. 植物学报 2(5), 52-54. |
| [2] | 商连光, 高振宇, 钱前 (2017). 作物杂种优势遗传基础的研究进展. 植物学报 52, 10-18. |
| [3] | 赵晶, 张弓 (2017). 翻译组学: 方法及应用. 生命的化学 37, 70-79. |
| [4] | Chotewutmontri P, Barkan A (2016). Dynamics of chloroplast translation during chloroplast differentiation in maize. PLoS Genet 12, e1006106. |
| [5] | Chotewutmontri P, Barkan A (2018). Multilevel effects of light on ribosome dynamics in chloroplasts program genome-wide and psbA-specific changes in translation. PLoS Genet 14, e1007555. |
| [6] | Gerashchenko MV, Gladyshev VN (2017). Ribonuclease selection for ribosome profiling. Nucleic Acids Res 45, e6. |
| [7] | Guo HW, Ecker JR (2003). Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell 115, 667-677. |
| [8] | Hsu PY, Calviello L, Wu HYL, Li FW, Rothfels CJ, Ohler U, Benfey PN (2016). Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proc Natl Acad Sci USA 113, E7126-E7135. |
| [9] | Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 7, 1534-1550. |
| [10] | Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218-223. |
| [11] | Ingolia NT, Lareau LF, Weissman JS (2011). Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789-802. |
| [12] | Jackson RJ, Hellen CUT, Pestova TV (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11, 113-127. |
| [13] | Juntawong P, Girke T, Bazin J, Bailey-Serres J (2014). Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci USA 111, E203-E212. |
| [14] | Konishi M, Yanagisawa S (2008). Two different mechanisms control ethylene sensitivity in Arabidopsis via the regulation of EBF2 expression. Plant Signal Behav 3, 749-751. |
| [15] | Kurihara Y, Makita Y, Kawashima M, Fujita T, Iwasaki S, Matsui M (2018). Transcripts from downstream alternative transcription start sites evade uORF-mediated inhibition of gene expression in Arabidopsis. Proc Natl Acad Sci USA 115, 7831-7836. |
| [16] | Kurihara Y, Makita Y, Shimohira H, Fujita T, Iwasaki S, Matsui M (2020). Translational landscape of protein- coding and non-protein-coding RNAs upon light exposure in Arabidopsis. Plant Cell Physiol 61, 536-545. |
| [17] | Lee S, Liu BT, Lee S, Huang SX, Shen B, Qian SB (2012). Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci USA 109, E2424-E2432. |
| [18] | Lei L, Shi JP, Chen J, Zhang M, Sun SL, Xie SJ, Li XJ, Zeng B, Peng LZ, Hauck A, Zhao HM, Song WB, Fan ZF, Lai JS (2015). Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress. Plant J 84, 1206-1218. |
| [19] | Li YR, Liu MJ (2020). Prevalence of alternative AUG and non-AUG translation initiators and their regulatory effects across plants. Genome Res 30, 1418-1433. |
| [20] | Liang Y, Zhu WC, Chen SJ, Qian JQ, Li L (2021). Genome-wide identification and characterization of small peptides in maize. Front Plant Sci 12, 695439. |
| [21] | Liu MJ, Wu SH, Wu JF, Lin WD, Wu YC, Tsai TY, Tsai HL, Wu SH (2013). Translational landscape of photomorphogenic Arabidopsis. Plant Cell 25, 3699-3710. |
| [22] | Lukoszek R, Feist P, Ignatova Z (2016). Insights into the adaptive response of Arabidopsis thaliana to prolonged thermal stress by ribosomal profiling and RNA-seq. BMC Plant Biol 16, 221. |
| [23] | Merchante C, Brumos J, Yun J, Hu QW, Spencer KR, Enríquez P, Binder BM, Heber S, Stepanova AN, Alonso JM (2015). Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell 163, 684-697. |
| [24] | Pajerowska-Mukhtar KM, Wang W, Tada Y, Oka N, Tucker CL, Fonseca JP, Dong XN (2012). The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr Biol 22, 103-112. |
| [25] | Planchard N, Bertin P, Quadrado M, Dargel-Graffin C, Hatin I, Namy O, Mireau H (2018). The translational landscape of Arabidopsis mitochondria. Nucleic Acids Res 46, 6218-6228. |
| [26] | Poidevin L, Forment J, Unal D, Ferrando A (2021). Transcriptome and translatome changes in germinated pollen under heat stress uncover roles of transporter genes involved in pollen tube growth. Plant Cell Environ 44, 2167-2184. |
| [27] | Schuster M, Gao Y, Sch?ttler MA, Bock R, Zoschke R (2020). Limited responsiveness of chloroplast gene expression during acclimation to high light in tobacco. Plant Physiol 182, 424-435. |
| [28] | Shamimuzzaman M, Vodkin L (2018). Ribosome profiling reveals changes in translational status of soybean transcripts during immature cotyledon development. PLoS One 13, e0194596. |
| [29] | Si XM, Zhang HW, Wang YP, Chen KL, Gao CX (2020). Manipulating gene translation in plants by CRISPR-Cas9- mediated genome editing of upstream open reading frames. Nat Protoc 15, 338-363. |
| [30] | Tr?sch R, Barahimipour R, Gao Y, Badillo-Corona JA, Gotsmann VL, Zimmer D, Mühlhaus T, Zoschke R, Willmund F (2018). Commonalities and differences of chloroplast translation in a green alga and land plants. Nat Plants 4, 564-575. |
| [31] | Tr?sch R, Ries F, Westrich LD, Gao Y, Herkt C, Hoppst?dter J, Heck-Roth J, Mustas M, Scheuring D, Choquet Y, R?schle M, Zoschke R, Willmund F (2022). Fast and global reorganization of the chloroplast protein biogenesis network during heat acclimation. Plant Cell 34, 1075-1099. |
| [32] | VanInsberghe M, van den Berg J, Andersson-Rolf A, Clevers H, van Oudenaarden A (2021). Single-cell Ribo- seq reveals cell cycle-dependent translational pausing. Nature 597, 561-565. |
| [33] | Wu HYL, Song GY, Walley JW, Hsu PY (2019). The tomato translational landscape revealed by transcriptome assembly and ribosome profiling. Plant Physiol 181, 367-380. |
| [34] | Wu LY, Lv YQ, Ye Y, Liang YR, Ye JH (2020). Transcriptomic and translatomic analyses reveal insights into the developmental regulation of secondary metabolism in the young shoots of tea plants (Camellia sinensis L.). J Agric Food Chem 68, 10750-10762. |
| [35] | Xing SN, Chen KL, Zhu HC, Zhang R, Zhang HW, Li BB, Gao CX (2020). Fine-tuning sugar content in strawberry. Genome Biol 21, 230. |
| [36] | Xiong QQ, Zhong L, Du J, Zhu CL, Peng XS, He XP, Fu JR, Ouyang LJ, Bian JM, Hu LF, Sun XT, Xu J, Zhou DH, Cai YC, Fu HH, He HH, Chen XR (2020). Ribosome profiling reveals the effects of nitrogen application translational regulation of yield recovery after abrupt drought- flood alternation in rice. Plant Physiol Biochem 155, 42-58. |
| [37] | Xu GY, Greene GH, Yoo H, Liu LJ, Marqués J, Motley J, Dong XN (2017a). Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature 545, 487-490. |
| [38] | Xu GY, Yuan M, Ai CR, Liu LJ, Zhuang E, Karapetyan S, Wang SP, Dong XN (2017b). uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 545, 491-494. |
| [39] | Xu QT, Liu Q, Chen ZT, Yue YP, Liu Y, Zhao Y, Zhou DX (2021). Histone deacetylases control lysine acetylation of ribosomal proteins in rice. Nucleic Acids Res 49, 4613-4628. |
| [40] | Yang XY, Song B, Cui J, Wang LN, Wang SS, Luo LL, Gao L, Mo BX, Yu Y, Liu L (2021). Comparative ribosome profiling reveals distinct translational landscapes of salt-sensitive and -tolerant rice. BMC Genomics 22, 612. |
| [41] | Yoo H, Greene GH, Yuan M, Xu GY, Burton D, Liu LJ, Marqués J, Dong XN (2020). Translational regulation of metabolic dynamics during effector-triggered immunity. Mol Plant 13, 88-98. |
| [42] | Zhang H, Wang YR, Lu J (2019). Function and evolution of upstream ORFs in eukaryotes. Trends Biochem Sci 44, 782-794. |
| [43] | Zhang HW, Si XM, Ji X, Fan R, Liu JX, Chen KL, Wang DW, Gao CX (2018). Genome editing of upstream open reading frames enables translational control in plants. Nat Biotechnol 36, 894-898. |
| [44] | Zhao H, Yin CC, Ma B, Chen SY, Zhang JS (2021). Ethylene signaling in rice and Arabidopsis: new regulators and mechanisms. J Integr Plant Biol 63, 102-125. |
| [45] | Zhao J, Qin B, Nikolay R, Spahn CMT, Zhang G (2019). Translatomics: the global view of translation. Int J Mol Sci 20, 212. |
| [46] | Zhu WC, Chen SJ, Zhang TF, Qian J, Luo Z, Zhao H, Zhang YR, Li L (2022). Dynamic patterns of the translatome in a hybrid triplet show translational fractionation of the maize subgenomes. Crop J 10, 36-46. |
| [47] | Zoschke R, Watkins KP, Barkan A (2013). A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell 25, 2265-2275. |