“后绿色革命”基因——助力培育“气候智能”作物新品种
- 中国农业大学生物学院, 北京 100193
收稿日期: 2025-02-11
录用日期: 2025-05-07
网络出版日期: 2025-05-14
基金资助
国家自然科学基金(32000216)
“Next-generation Green Revolution” Genes: Toward New “Climate-Smart” Crop Breeding
- College of Biological Sciences, China Agricultural University, Beijing 100193, China
Received date: 2025-02-11
Accepted date: 2025-05-07
Online published: 2025-05-14
摘要
本文引用格式
马亮 , 杨永青 , 郭岩 . “后绿色革命”基因——助力培育“气候智能”作物新品种[J]. 植物学报, 2025 , 60(4) : 489 -498 . DOI: 10.11983/CBB25021
Abstract
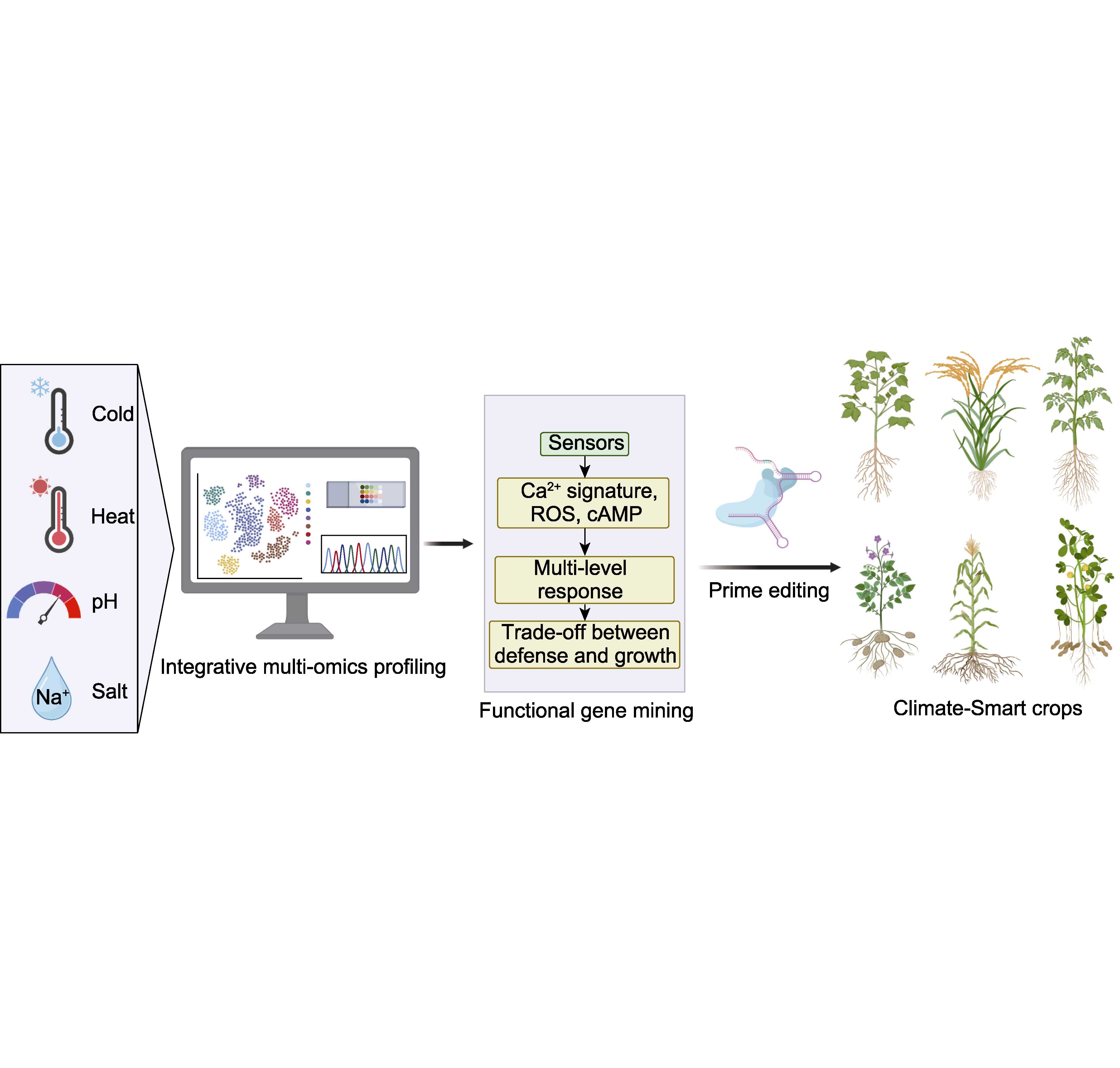
In recent years, significant progresses have been made in plant stress biology, particularly in elucidating the mechanisms underlying responses to extreme temperatures and salinity-alkalinity stresses. These advancements have not only broadened our understanding of plant resilience but also provided a wealth of potential targets for molecular breeding, paving new avenues for developing climate-resilient crop varieties that maintain high yield potential under both optimal and adverse conditions. This review concisely summarizes current knowledge on signal perception and transduction mechanisms during plant adaptation to extreme temperature and saline-alkali stresses, discusses the balance regulation between growth/development and stress tolerance, particularly highlights recent breakthroughs by Chinese scientists in discovering key genes and deciphering mechanisms that synergistically improve crop stress resistance and yield. We also provide future prospects for breeding strategies.
参考文献
| [1] | Allen DJ, Ort DR (2001). Impacts of chilling temperatures on photosynthesis in warm-climate plants. Trends Plant Sci 6, 36-42. |
| [2] | Banani SF, Lee HO, Hyman AA, Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18, 285-298. |
| [3] | Bohn L, Huang J, Weidig S, Yang ZY, Heidersberger C, Genty B, Falter-Braun P, Christmann A, Grill E (2024). The temperature sensor TWA1 is required for thermotolerance in Arabidopsis. Nature 629, 1126-1132. |
| [4] | Chen K, Gao JH, Sun SJ, Zhang ZJ, Yu B, Li J, Xie CG, Li GJ, Wang PC, Song CP, Bressan RA, Hua J, Zhu JK, Zhao Y (2020). BONZAI proteins control global osmotic stress responses in plants. Curr Biol 30, 4815-4825. |
| [5] | Chen LP, Zhao Y, Xu SJ, Zhang ZY, Xu YY, Zhang JY, Chong K (2018). OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytol 218, 219-231. |
| [6] | Choi WG, Toyota M, Kim SH, Hilleary R, Gilroy S (2014). Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc Natl Acad Sci USA 111, 6497-6502. |
| [7] | Chung BYW, Balcerowicz M, Di Antonio M, Jaeger KE, Geng F, Franaszek K, Marriott P, Brierley I, Firth AE, Wigge PA (2020). An RNA thermoswitch regulates daytime growth in Arabidopsis. Nat Plants 6, 522-532. |
| [8] | Ding YL, Shi YT, Yang SH (2024). Regulatory networks underlying plant responses and adaptation to cold stress. Annu Rev Genet 58, 43-65. |
| [9] | Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21, 972-984. |
| [10] | Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, Duan QH, Liu MC, Maman J, Steinhorst L, Schmitz- Thom I, Yvon R, Kudla J, Wu HM, Cheung AY, Dinneny JR (2018). The FERONIA receptor kinase maintains cell- wall integrity during salt stress through Ca2+ signaling. Curr Biol 28, 666-675. |
| [11] | Gao L, Jiang HF, Li MZ, Wang DF, Xiang HT, Zeng R, Chen LM, Zhang XY, Zuo JR, Yang SH, Shi YT (2024a). Genetic and lipidomic analyses reveal the key role of lipid metabolism for cold tolerance in maize. J Genet Genomics 51, 326-337 |
| [12] | Gao L, Pan LL, Shi YT, Zeng R, Li MZ, Li ZY, Zhang X, Zhao XM, Gong XR, Huang W, Yang XH, Lai JS, Zuo JR, Gong ZZ, Wang XQ, Jin WW, Dong ZB, Yang SH (2024b). Genetic variation in a heat shock transcription factor modulates cold tolerance in maize. Mol Plant 17, 1423-1438. |
| [13] | Guo SQ, Chen YX, Ju YL, Pan CY, Shan JX, Ye WW, Dong NQ, Kan Y, Yang YB, Zhao HY, Yu HX, Lu ZQ, Lei JJ, Liao B, Mu XR, Cao YJ, Guo LX, Gao J, Zhou JF, Yang KY, Lin HX, Lin YS (2025). Fine-tuning gibberellin improves rice alkali-thermal tolerance and yield. Nature 639, 162-171. |
| [14] | Guo SY, Xu YY, Liu HH, Mao ZW, Zhang C, Ma Y, Zhang QR, Meng Z, Chong K (2013). The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF- 14. Nat Commun 4, 1566. |
| [15] | Guo XY, Zhang DJ, Wang ZL, Xu SJ, Batisti? O, Steinhorst L, Li H, Weng YX, Ren DT, Kudla J, Xu YY, Chong K (2023). Cold-induced calreticulin OsCRT3 conformational changes promote OsCIPK7 binding and temperature sensing in rice. EMBO J 42, e110518. |
| [16] | Jiang HF, Shi YT, Liu JY, Li Z, Fu DY, Wu SF, Li MZ, Yang ZJ, Shi YL, Lai JS, Yang XH, Gong ZZ, Hua J, Yang SH (2022). Natural polymorphism of ZmICE1 contributes to amino acid metabolism that impacts cold tolerance in maize. Nat Plants 8, 1176-1190. |
| [17] | Jiang ZH, Zhou XP, Tao M, Yuan F, Liu LL, Wu FH, Wu XM, Xiang Y, Niu Y, Liu F, Li CJ, Ye R, Byeon B, Xue Y, Zhao HY, Wang HN, Crawford BM, Johnson DM, Hu CX, Pei C, Zhou WM, Swift GB, Zhang H, Vo-Dinh T, Hu ZL, Siedow JN, Pei ZM (2019). Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 572, 341-346. |
| [18] | Jung JH, Barbosa AD, Hutin S, Kumita JR, Gao MJ, Derwort D, Silva CS, Lai XL, Pierre E, Geng F, Kim SB, Baek S, Zubieta C, Jaeger KE, Wigge PA (2020). A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 585, 256-260. |
| [19] | Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao MJ, Khattak AK, Box MS, Charoensawan V, Cortijo S, Kumar M, Grant A, Locke JCW, Sch?fer E, Jaeger KE, Wigge PA (2016). Phytochromes function as thermosensors in Arabidopsis. Science 354, 886-889. |
| [20] | Kan Y, Mu XR, Zhang H, Gao J, Shan JX, Ye WW, Lin HX (2022). TT2 controls rice thermotolerance through SCT1- dependent alteration of wax biosynthesis. Nat Plants 8, 53-67. |
| [21] | Kiegle E, Moore CA, Haseloff J, Tester MA, Knight MR (2000). Cell-type-specific calcium responses to drought, salt and cold in the Arabidopsis root. Plant J 23, 267-278. |
| [22] | Kim Y, Park S, Gilmour SJ, Thomashow MF (2013). Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J 75, 364-376. |
| [23] | Knight H, Trewavas AJ, Knight MR (1997). Calcium signaling in Arabidopsis thaliana responding to drought and salinity. Plant J 12, 1067-1078. |
| [24] | Laohavisit A, Richards SL, Shabala L, Chen C, Cola?o RDDR, Swarbreck SM, Shaw E, Dark A, Shabala S, Shang ZL, Davies JM (2013). Salinity-induced calcium signaling and root adaptation in Arabidopsis require the calcium regulatory protein annexin1. Plant Physiol 163, 253-262. |
| [25] | Legris M, Klose C, Burgie ES, Rojas CCB, Neme M, Hiltbrunner A, Wigge PA, Sch?fer E, Vierstra RD, Casal JJ (2016). Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354, 897-900. |
| [26] | Li ZY, Fu DY, Wang X, Zeng R, Zhang X, Tian JG, Zhang SS, Yang XH, Tian F, Lai JS, Shi YT, Yang SH (2022). The transcription factor bZIP68 negatively regulates cold tolerance in maize. Plant Cell 34, 2833-2851. |
| [27] | Liu ZY, Jia YX, Ding YL, Shi YT, Li Z, Guo Y, Gong ZZ, Yang SH (2017). Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol Cell 66, 117-128. |
| [28] | Lou HC, Li SJ, Shi ZH, Zou YP, Zhang YQ, Huang XZ, Yang DD, Yang YF, Li ZY, Xu C (2025). Engineering source-sink relations by prime editing confers heat-stress resilience in tomato and rice. Cell 188, 530-549. |
| [29] | Luo W, Xu YY, Cao J, Guo XY, Han JD, Zhang YY, Niu YD, Zhang ML, Wang Y, Liang GH, Qian Q, Ge S, Chong K (2024). COLD6-OSM1 module senses chilling for cold tolerance via 2',3'-cAMP signaling in rice. Mol Cell 84, 4224-4238. |
| [30] | Ma L, Liu XH, Lv WJ, Yang YQ (2022). Molecular mechanisms of plant responses to salt stress. Front Plant Sci 13, 934877. |
| [31] | Ma L, Ye JM, Yang YQ, Lin HX, Yue LL, Luo J, Long Y, Fu HQ, Liu XN, Zhang YL, Wang Y, Chen LY, Kudla J, Wang YJ, Han SC, Song CP, Guo Y (2019). The SOS2-SCaBP8 complex generates and fine-tunes an AtANN4-dependent calcium signature under salt stress. Dev Cell 48, 697-709. |
| [32] | Ma Y, Dai XY, Xu YY, Luo W, Zheng XM, Zeng DL, Pan YJ, Lin XL, Liu HH, Zhang DJ, Xiao J, Guo XY, Xu SJ, Niu YD, Jin JB, Zhang H, Xu X, Li LG, Wang W, Qian Q, Ge S, Chong K (2015). COLD1 confers chilling tolerance in rice. Cell 160, 1209-1221. |
| [33] | Mason TG, Maskell EJ (1928). Studies on the transport of carbohydrates in the cotton plant: II. The factors determining the rate and the direction of movement of sugars. Ann Bot 42, 571-636. |
| [34] | Matsuoka Y, Vigouroux Y, Goodman MM, Sanchez GJ, Buckler E, Doebley J (2002). A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad Sci USA 99, 6080-6084. |
| [35] | Reindl A, Sch?ffl F, Schell J, Koncz C, Bakó L (1997). Phosphorylation by a cyclin-dependent kinase modulates DNA binding of the Arabidopsis heat-shock transcription factor HSF1 in vitro. Plant Physiol 115, 93-100. |
| [36] | Steinhorst L, He GF, Moore LK, Schültke S, Schmitz- Thom I, Cao YB, Hashimoto K, Andrés Z, Piepenburg K, Ragel P, Behera S, Almutairi BO, Batisti? O, Wyganowski T, K?ster P, Edel KH, Zhang CX, Krebs M, Jiang CF, Guo Y, Quintero FJ, Bock R, Kudla J (2022). A Ca2+-sensor switch for tolerance to elevated salt stress in Arabidopsis. Dev Cell 57, 2081-2094. |
| [37] | Tracy FE, Gilliham M, Dodd AN, Webb AAR, Tester M (2008). NaCl-induced changes in cytosolic free Ca2+ in Arabidopsis thaliana are heterogeneous and modified by external ionic composition. Plant Cell Environ 31, 1063-1073. |
| [38] | van Dijk M, Morley T, Rau ML, Saghai Y (2021). A meta- analysis of projected global food demand and population at risk of hunger for the period 2010-2050. Nat Food 2, 494-501. |
| [39] | Wang BY, Zhang HH, Huai JL, Peng FY, Wu J, Lin RC, Fang XF (2022). Condensation of SEUSS promotes hyperosmotic stress tolerance in Arabidopsis. Nat Chem Biol 18, 1361-1369. |
| [40] | Wang XH, Zhao C, Müller C, Wang CZ, Ciais P, Janssens I, Pe?uelas J, Asseng S, Li T, Elliott J, Huang Y, Li L, Piao S (2020). Emergent constraint on crop yield response to warmer temperature from field experiments. Nat Sustain 3, 908-916. |
| [41] | Wang ZY, Yang QH, Zhang D, Lu YY, Wang YC, Pan YJ, Qiu YP, Men Y, Yan W, Xiao ZN, Sun RX, Li WY, Huang HD, Guo HW (2024). A cytoplasmic osmosensing mechanism mediated by molecular crowding-sensitive DCP5. Science 386, eadk9067. |
| [42] | Wheeler T, Von Braun J (2013). Climate change impacts on global food security. Science 341, 508-513. |
| [43] | Wu Y, Wang Y, Mi XF, Shan JX, Li XM, Xu JL, Lin HX (2016). The QTL GNP1 encodes GA20ox1, which increases grain number and yield by increasing cytokinin activity in rice panicle meristems. PLoS Genet 12, e1006386. |
| [44] | Xia CX, Liang GH, Chong K, Xu YY (2023). The COG1- OsSERL2 complex senses cold to trigger signaling network for chilling tolerance in japonica rice. Nat Commun 14, 3104. |
| [45] | Xiang YH, Yu JJ, Liao B, Shan JX, Ye WW, Dong NQ, Guo T, Kan Y, Zhang H, Yang YB, Li YC, Zhao HY, Yu HX, Lu ZQ, Lin HX (2022). An α/β hydrolase family member negatively regulates salt tolerance but promotes flowering through three distinct functions in rice. Mol Plant 15, 1908-1930. |
| [46] | Yang YQ, Guo Y (2018). Unraveling salt stress signaling in plants. J Integr Plant Biol 60, 796-804. |
| [47] | Yang ZR, Cao YB, Shi YT, Qin F, Jiang CF, Yang SH (2023). Genetic and molecular exploration of maize environmental stress resilience: toward sustainable agriculture. Mol Plant 16, 1496-1517. |
| [48] | Yin WC, Xiao YH, Niu M, Meng WJ, Li LL, Zhang XX, Liu DP, Zhang GX, Qian YW, Sun ZT, Huang RY, Wang SP, Liu CM, Chu CC, Tong HN (2020). ARGONAUTE2 enhances grain length and salt tolerance by activating BIG GRAIN3 to modulate cytokinin distribution in rice. Plant Cell 32, 2292-2306. |
| [49] | Yuan F, Yang HM, Xue Y, Kong DD, Ye R, Li CJ, Zhang JY, Theprungsirikul L, Shrift T, Krichilsky B, Johnson DM, Swift GB, He YK, Siedow JN, Pei ZM (2014). OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 514, 367-371. |
| [50] | Zeng R, Li ZY, Shi YT, Fu DY, Yin P, Cheng JK, Jiang CF, Yang SH (2021). Natural variation in a type-A response regulator confers maize chilling tolerance. Nat Commun 12, 4713. |
| [51] | Zeng R, Shi YT, Guo L, Fu DY, Li MZ, Zhang XY, Li ZY, Zhuang JH, Yang XH, Zuo JR, Gong ZZ, Tian F, Yang SH (2025). A natural variant of COOL1 gene enhances cold tolerance for high-latitude adaptation in maize. Cell 188, 1315-1329. |
| [52] | Zhang DJ, Guo XY, Xu YY, Li H, Ma L, Yao XF, Weng YX, Guo Y, Liu CM, Chong K (2019). OsCIPK7 point-mutation leads to conformation and kinase-activity change for sensing cold response. J Integr Plant Biol 61, 1194-1200. |
| [53] | Zhang H, Zhang JY, Xu QY, Wang DD, Di H, Huang J, Yang XW, Wang ZF, Zhang L, Dong L, Wang ZH, Zhou Y (2020). Identification of candidate tolerance genes to low-temperature during maize germination by GWAS and RNA-seq approaches. BMC Plant Biol 20, 333. |
| [54] | Zhang H, Zhou JF, Kan Y, Shan JX, Ye WW, Dong NQ, Guo T, Xiang YH, Yang YB, Li YC, Zhao HY, Yu HX, Lu ZQ, Guo SQ, Lei JJ, Liao B, Mu XR, Cao YJ, Yu JJ, Lin YS, Lin HX (2022). A genetic module at one locus in rice protects chloroplasts to enhance thermotolerance. Science 376, 1293-1300. |
| [55] | Zhou XM, Muhammad I, Lan H, Xia C (2022). Recent advances in the analysis of cold tolerance in maize. Front Plant Sci 13, 866034. |