

Chinese Bulletin of Botany ›› 2017, Vol. 52 ›› Issue (6): 723-732.DOI: 10.11983/CBB16237
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Song Huifang, Liu Haishuang, Yang Yiming, Fan Shutian, Li Changyu*( ), Ai Jun*(
), Ai Jun*( )
)
Received:2016-12-02
Accepted:2017-01-24
Online:2017-11-01
Published:2018-02-22
Contact:
Li Changyu,Ai Jun
Song Huifang, Liu Haishuang, Yang Yiming, Fan Shutian, Li Changyu, Ai Jun. Screening of Universal DNA Barcodes for Vitis amurensis[J]. Chinese Bulletin of Botany, 2017, 52(6): 723-732.
| Number | Varieties name | Locality of origin | Parents or source | Flower type |
|---|---|---|---|---|
| 1 | Zuoyouhong | Zuojia, Jilin | Varieties | Bisexual |
| 2 | Shuanghong | Zuojia, Jilin | Varieties | Bisexual |
| 3 | Zuoshan1 | Zuojia, Jilin | Wild resource | Male |
| 4 | Zuoshan2 | Zuojia, Jilin | Wild resource | Male |
| 5 | 4N1 | Zuojia, Jilin | Genetic material | Tetraploid |
| 6 | 4N2 | Zuojia, Jilin | Genetic material | Tetraploid |
| 7 | Shuangqing | Zuojia, Jilin | Varieties | Bisexual |
| 8 | Shuangfeng | Zuojia, Jilin | Varieties | Bisexual |
| 9 | Shuangyou | Ji’an, Jilin | Varieties | Bisexual |
| 10 | 75047 | Shangzhi, Heilongjiang | Wild resource | Female |
| 11 | 73061 | Dunhua, Jilin | Wild resource | Female |
Table 1 Information of Vitis amurensis varieties
| Number | Varieties name | Locality of origin | Parents or source | Flower type |
|---|---|---|---|---|
| 1 | Zuoyouhong | Zuojia, Jilin | Varieties | Bisexual |
| 2 | Shuanghong | Zuojia, Jilin | Varieties | Bisexual |
| 3 | Zuoshan1 | Zuojia, Jilin | Wild resource | Male |
| 4 | Zuoshan2 | Zuojia, Jilin | Wild resource | Male |
| 5 | 4N1 | Zuojia, Jilin | Genetic material | Tetraploid |
| 6 | 4N2 | Zuojia, Jilin | Genetic material | Tetraploid |
| 7 | Shuangqing | Zuojia, Jilin | Varieties | Bisexual |
| 8 | Shuangfeng | Zuojia, Jilin | Varieties | Bisexual |
| 9 | Shuangyou | Ji’an, Jilin | Varieties | Bisexual |
| 10 | 75047 | Shangzhi, Heilongjiang | Wild resource | Female |
| 11 | 73061 | Dunhua, Jilin | Wild resource | Female |
| Fragment | Eight annealing temperature gradient (°C) | Annealing temperature (°C) |
|---|---|---|
| ITS2 | 54.8-55.4-56.0-56.6-57.2-57.8-58.4-59.0 | 56.0 |
| psbA-trnH | 52.2-52.8-53.4-54.0-54.6-55.2-55.8-56.4 | 55.2 |
| matK | 48.9-49.5-50.1-50.7-51.3-51.9-52.5-53.1 | 50.1 |
| rbcL | 52.1-52.7-53.3-53.9-54.5-55.1-55.7-56.3 | 54.5 |
| ITS | 49.9-50.5-51.1-51.7-52.3-52.9-53.5-54.1 | 52.9 |
Table 2 The primer information and annealing temperature of PCR from Vitis amurensis
| Fragment | Eight annealing temperature gradient (°C) | Annealing temperature (°C) |
|---|---|---|
| ITS2 | 54.8-55.4-56.0-56.6-57.2-57.8-58.4-59.0 | 56.0 |
| psbA-trnH | 52.2-52.8-53.4-54.0-54.6-55.2-55.8-56.4 | 55.2 |
| matK | 48.9-49.5-50.1-50.7-51.3-51.9-52.5-53.1 | 50.1 |
| rbcL | 52.1-52.7-53.3-53.9-54.5-55.1-55.7-56.3 | 54.5 |
| ITS | 49.9-50.5-51.1-51.7-52.3-52.9-53.5-54.1 | 52.9 |
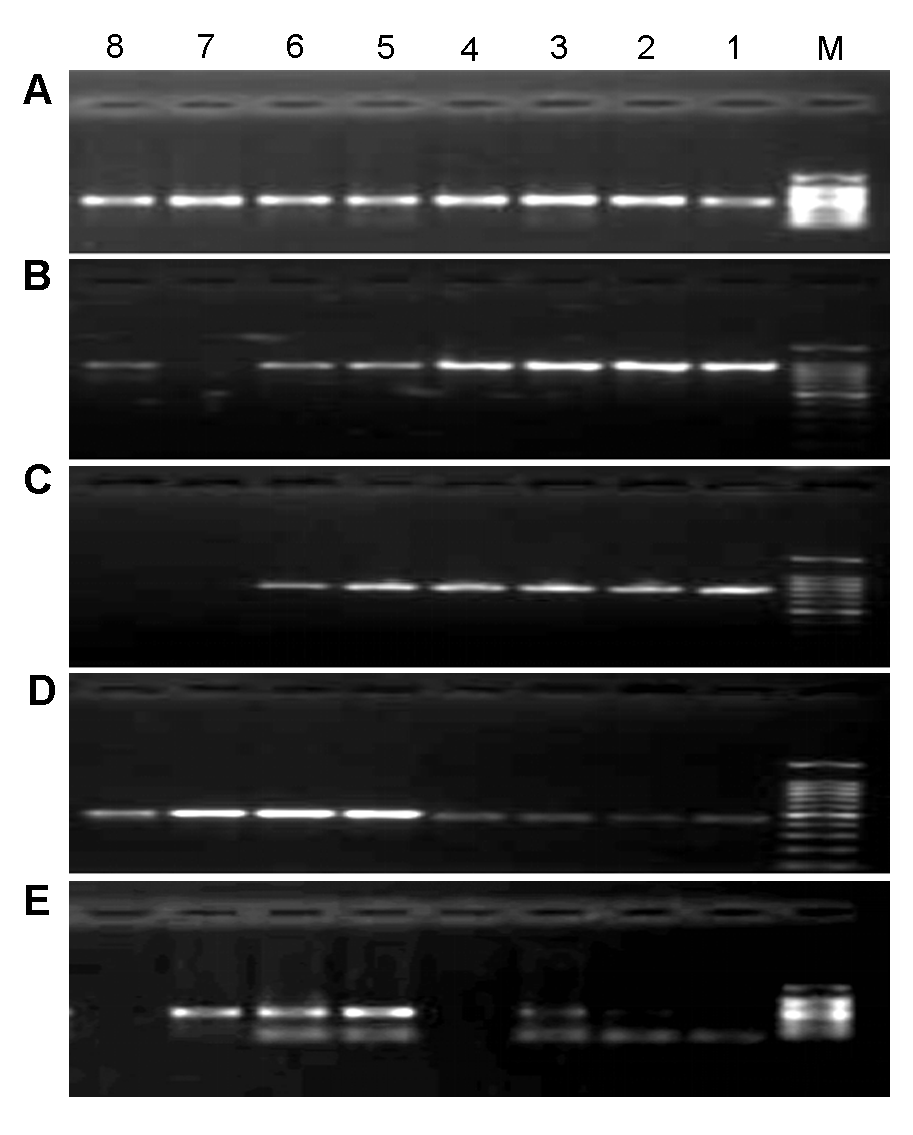
Figure 1 Gradient annealing temperature of PCR reaction of 5 candidate sequences(A) ITS2; (B) matK; (C) rbcL; (D) psbA-trnH; (E) ITS. M: DNA marker DL2000; 1-8: Gradient annealing temperature among 45-60°C
| Marker | Amplification efficiency (%) | Sequencing success rate (%) | Effective sequence ratio (%) |
|---|---|---|---|
| ITS2 | 100.0 | 96.9 | 96.9 |
| matK | 100.0 | 96.9 | 96.9 |
| psbA-trnH | 100.0 | 100.0 | 100.0 |
| rbcL | 96.9 | 90.9 | 88.1 |
| ITS | 45.5 | 30.3 | 13.8 |
Table 3 The effective sequence ratio obtained by PCR am- plification of five DNA barcode sequences
| Marker | Amplification efficiency (%) | Sequencing success rate (%) | Effective sequence ratio (%) |
|---|---|---|---|
| ITS2 | 100.0 | 96.9 | 96.9 |
| matK | 100.0 | 96.9 | 96.9 |
| psbA-trnH | 100.0 | 100.0 | 100.0 |
| rbcL | 96.9 | 90.9 | 88.1 |
| ITS | 45.5 | 30.3 | 13.8 |
| Potential barcode | Aligned length (bp) | Number of variable sites | Mean intra- distance | Mean inter- distance | Average of GC content (%) |
|---|---|---|---|---|---|
| ITS2 | 483 | 397 | 0.0015 | 0.1162 | 64.50 |
| matK | 896 | 44 | 0.0032 | 0.0110 | 35.40 |
| psbA-trnH | 422 | 238 | 0.0089 | 0.0921 | 27.50 |
| rbcL | 697 | 82 | 0.0068 | 0.0180 | 44.20 |
| ITS2+matK | 1379 | 441 | 0.0024 | 0.0652 | 49.95 |
| ITS2+psbA-trnH | 905 | 635 | 0.0058 | 0.0985 | 46.00 |
| ITS2+rbcL | 1180 | 479 | 0.0045 | 0.0655 | 54.35 |
| matK+psbA-trnH | 1318 | 282 | 0.0064 | 0.0550 | 31.45 |
| matK+rbcL | 1543 | 126 | 0.0051 | 0.0220 | 41.35 |
| psbA-trnH+rbcL | 1119 | 320 | 0.0076 | 0.0560 | 39.80 |
Table 4 Measures of inter-varieties and intra-varieties divergence locus length and average of GC content for 4 candidate barcodes/combination sequences
| Potential barcode | Aligned length (bp) | Number of variable sites | Mean intra- distance | Mean inter- distance | Average of GC content (%) |
|---|---|---|---|---|---|
| ITS2 | 483 | 397 | 0.0015 | 0.1162 | 64.50 |
| matK | 896 | 44 | 0.0032 | 0.0110 | 35.40 |
| psbA-trnH | 422 | 238 | 0.0089 | 0.0921 | 27.50 |
| rbcL | 697 | 82 | 0.0068 | 0.0180 | 44.20 |
| ITS2+matK | 1379 | 441 | 0.0024 | 0.0652 | 49.95 |
| ITS2+psbA-trnH | 905 | 635 | 0.0058 | 0.0985 | 46.00 |
| ITS2+rbcL | 1180 | 479 | 0.0045 | 0.0655 | 54.35 |
| matK+psbA-trnH | 1318 | 282 | 0.0064 | 0.0550 | 31.45 |
| matK+rbcL | 1543 | 126 | 0.0051 | 0.0220 | 41.35 |
| psbA-trnH+rbcL | 1119 | 320 | 0.0076 | 0.0560 | 39.80 |
| w+ | w- | Inter relative ranks | n | P value | Result |
|---|---|---|---|---|---|
| ITS2 | rbcL | w+=33035.00, w-=13630.00 | 319 | 0.000 | P<0.01, ITS2>rbcL |
| ITS2 | matK | w+=1444453.00, w-=0.00 | 577 | 0.000 | P<0.01, ITS2>matK |
| ITS2 | psbA-trnH | w+=11562.00, w-=8660.00 | 500 | 0.000 | P<0.01, ITS2>psbA-trnH |
| rbcL | matK | w+=8673.00, w-=6903.00 | 319 | 0.174 | P>0.05, rbcL=matK |
| rbcL | psbA-trnH | w+=4350.00, w-=8720.00 | 620 | 0.000 | P<0.01, rbcL<psbA-trnH |
| matK | psbA-trnH | w+=12556.00, w-=25330.00 | 422 | 0.000 | P<0.01, matK<psbA-trnH |
Table 5 Wilcoxon signes tests for inter-varieties divergences of candidate sequences
| w+ | w- | Inter relative ranks | n | P value | Result |
|---|---|---|---|---|---|
| ITS2 | rbcL | w+=33035.00, w-=13630.00 | 319 | 0.000 | P<0.01, ITS2>rbcL |
| ITS2 | matK | w+=1444453.00, w-=0.00 | 577 | 0.000 | P<0.01, ITS2>matK |
| ITS2 | psbA-trnH | w+=11562.00, w-=8660.00 | 500 | 0.000 | P<0.01, ITS2>psbA-trnH |
| rbcL | matK | w+=8673.00, w-=6903.00 | 319 | 0.174 | P>0.05, rbcL=matK |
| rbcL | psbA-trnH | w+=4350.00, w-=8720.00 | 620 | 0.000 | P<0.01, rbcL<psbA-trnH |
| matK | psbA-trnH | w+=12556.00, w-=25330.00 | 422 | 0.000 | P<0.01, matK<psbA-trnH |
| w+ | w- | Inter relative ranks | n | P value | Result |
|---|---|---|---|---|---|
| ITS2 | rbcL | w+=39345.00, w-=18966.00 | 384 | 0.000 | P<0.01, ITS2>rbcL |
| ITS2 | matK | w+=142845.00, w-=0.00 | 577 | 0.000 | P<0.01, ITS2>matK |
| ITS2 | psbA-trnH | w+=17550.00, w-=18326.00 | 366 | 0.056 | P>0.05, ITS2=psbA-trnH |
| rbcL | matK | w+=11952.00, w-=5253.00 | 384 | 0.000 | P<0.01, rbcL>matK |
| rbcL | psbA-trnH | w+=5968.00, w-=13589.00 | 469 | 0.000 | P<0.01, rbc<psbA-trnH |
| matK | psbA-trnH | w+=3985.00, w-=12578.00 | 580 | 0.000 | P<0.01, matK<psbA-trnH |
Table 6 Wilcoxon signes tests for intra-varieties divergences of candidate sequences
| w+ | w- | Inter relative ranks | n | P value | Result |
|---|---|---|---|---|---|
| ITS2 | rbcL | w+=39345.00, w-=18966.00 | 384 | 0.000 | P<0.01, ITS2>rbcL |
| ITS2 | matK | w+=142845.00, w-=0.00 | 577 | 0.000 | P<0.01, ITS2>matK |
| ITS2 | psbA-trnH | w+=17550.00, w-=18326.00 | 366 | 0.056 | P>0.05, ITS2=psbA-trnH |
| rbcL | matK | w+=11952.00, w-=5253.00 | 384 | 0.000 | P<0.01, rbcL>matK |
| rbcL | psbA-trnH | w+=5968.00, w-=13589.00 | 469 | 0.000 | P<0.01, rbc<psbA-trnH |
| matK | psbA-trnH | w+=3985.00, w-=12578.00 | 580 | 0.000 | P<0.01, matK<psbA-trnH |
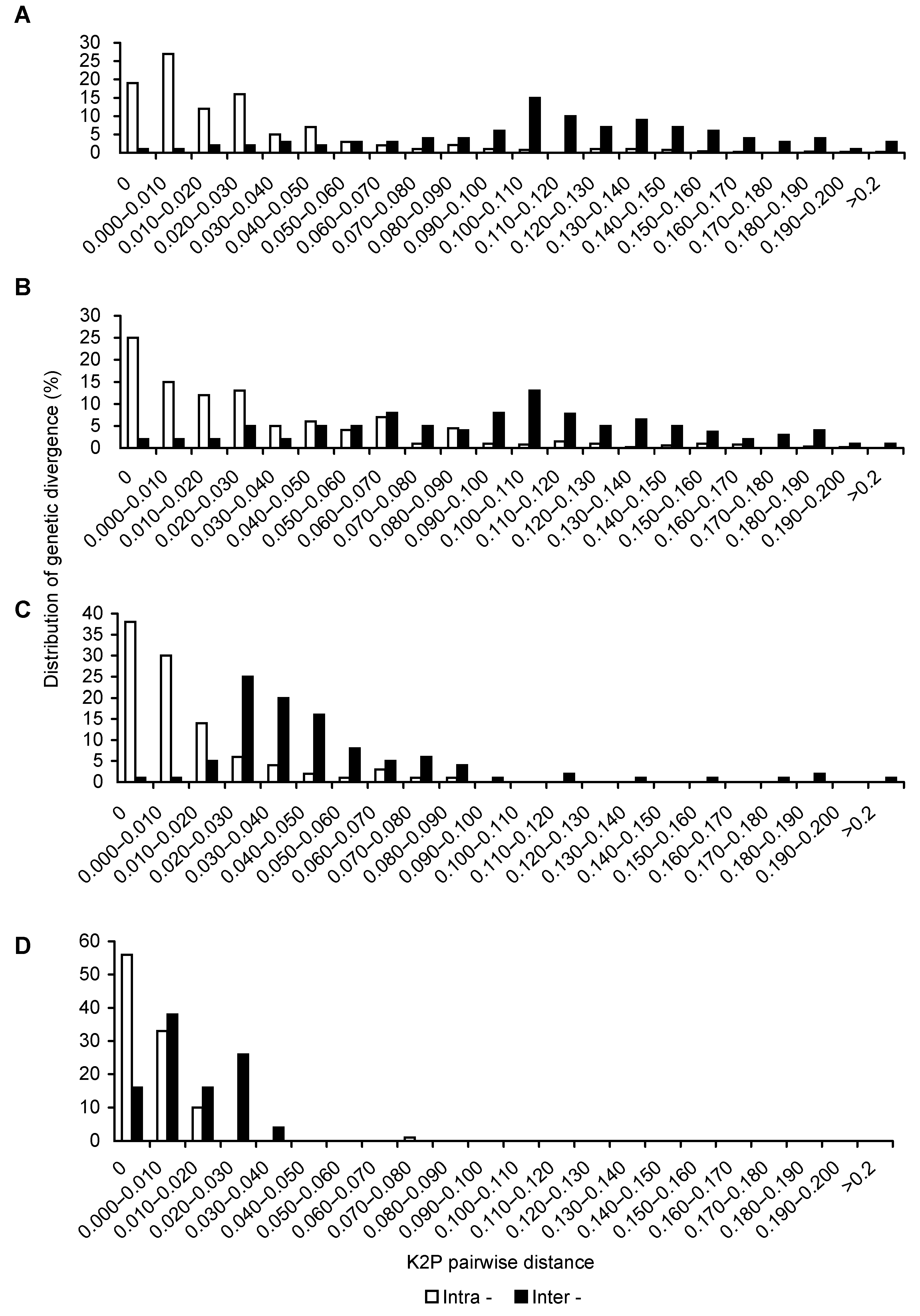
Figure 2 Distribution for intra- and inter-varieties variation of Vitis amurensis(A) ITS2 sequence; (B) psbA-trnH sequence; (C) rbcL sequence; (D) matK sequence
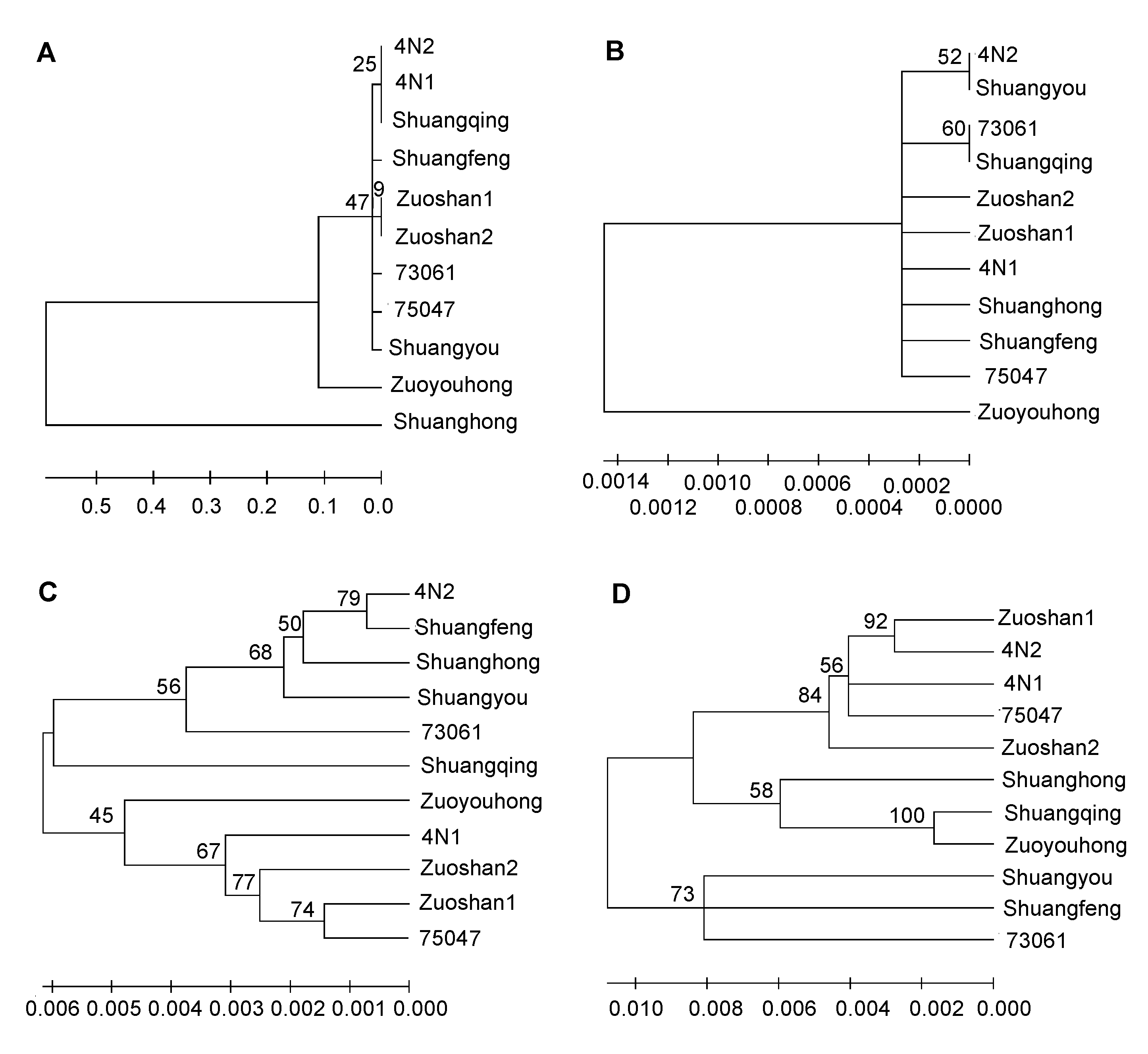
Figure 3 Neighbor-joining (NJ) tree for 11 varieties of Vitis amurensis by using different sequences(A) ITS2 sequence; (B) psbA-trnH sequence; (C) rbcL sequence; (D) matK sequence
| [1] |
陈士林, 姚辉, 韩建萍, 辛天怡, 庞晓慧, 石林春, 罗焜, 宋经元, 侯典云, 石上梅, 钱忠直 (2013). 中药材DNA条形码分子鉴定指导原则. 中国中药杂志 38, 141-148.
DOI URL |
| [2] |
东秀珠, 沈德龙, 辛玉华 (2000). 16S rDNA同源性所揭示的双歧杆菌与有关细菌的亲缘关系. 生物多样性 8, 146-152.
DOI URL |
| [3] |
高健, 孟婉姮, 杜芳, 李俊清 (2015). 鸡爪槭种下分类群的DNA条形码筛选. 植物科学学报33, 734-743.
DOI URL |
| [4] |
李晓艳, 杨义明, 范书田, 王振兴, 艾军, 沈育杰 (2014). 山葡萄种质资源收集、保存、评价与利用研究进展. 河北林业科技 (5-6), 115-121.
DOI URL |
| [5] |
刘震, 陈科力, 罗焜, 潘宏林, 陈士林 (2010). 忍冬科药用植物DNA条形码通用序列的筛选. 中国中药杂志 35, 2527-2532.
DOI URL |
| [6] | 任保青, 陈之端 (2010). 植物DNA条形码技术. 植物学报45, 1-12. |
| [7] |
任阳阳, 张梦婷, 张嘉丽, 樊佳佳, 张晓存, 王俊, 刘霞 (2016). 虾脊兰属植物DNA条形码的确立. 世界中医药 11, 2425-2429.
DOI URL |
| [8] |
沈育杰, 赵淑兰, 杨义明, 李晓红, 宋润刚, 路文鹏 (2006). 我国山葡萄种质资源研究与利用现状. 特产研究28(3), 53-57.
DOI URL |
| [9] |
石志刚, 万如, 李彦龙, 王亚军, 马婷慧 (2016). 宁夏枸杞主要品种psbA-trnH的DNA条形码鉴定的初步研究. 农业科技与装备(6), 1-2, 7.
DOI URL |
| [10] |
宋润刚, 艾军, 李晓红, 杨义明, 沈育杰 (2009). 中国山葡萄产业的发展及对策. 中外葡萄与葡萄酒 (11), 64-69.
DOI URL |
| [11] |
王柯, 陈科力, 刘震, 陈士林 (2011). 锦葵科植物DNA条形码通用序列的筛选. 植物学报 46, 276-284.
DOI URL |
| [12] | 辛天怡, 姚辉, 罗焜, 向丽, 马晓冲, 韩建萍, 林余霖, 宋经元, 陈士林 (2012). 羌活药材ITS/ITS2条形码鉴定及其稳定性与准确性研究. 药学学报 47, 1098-1105. |
| [13] |
CBOL Plant Working Group (2009). A DNA barcode for land plants.Proc Natl Acad Sci USA 106, 12794-12797.
DOI URL PMID |
| [14] |
Chase MW, Cowan RS, Hollingsworth PM, van den Berg C, Madri?án S, Petersen G, Seberg O, J?rgsensen T, Cameron KM, Carine M, Pedersen N, Hedderson TAJ, Conrad F, Salazar GA, Richardson JE, Hollingsworth ML, Barraclough TG, Kelly L, Wilkinson M (2007). A proposal for a standardised protocol to barcode all land plants.Taxon 56, 295-299.
DOI URL |
| [15] |
Chen SL, Yao H, Han JP, Liu C, Song JY, Shi LC, Zhu YJ, Ma XY, Gao T, Pang XH, Luo K, Li Y, Li XW, Jia XC, Lin YL, Leon C (2010). Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species.PLoS One 5, e8613.
DOI URL PMID |
| [16] |
China Plant BOL Group, Li DZ, Gao LM, Li HT, Wang H, Ge XJ, Liu JQ, Chen ZD, Zhou SL, Chen SL, Yang JB, Fu CX, Zeng CX, Yan HF, Zhu YJ, Sun YS, Chen SY, Zhao L, Wang K, Yang T, Duan GW (2011). Comparative analysis of a large dataset indicates that internal trans- cribed spacer (ITS) should be incorporated into the core barcode for seed plants.Proc Natl Acad Sci USA 108, 19641-19646.
DOI URL PMID |
| [17] | Dassanayake RS, Gunawardene YINS, De Silva BDDNK (2008). ITS-2 secondary structures and phylogeny of Ano- pheles culicifacies species. Bioinformation 2, 456-460. |
| [18] |
Fazekas AJ, Burgess KS, Kesanakurti PR, Graham SW, Newmaster SG, Husband BC, Percy DM, Hajibabaei M, Barrett SCH (2008). Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well.PLoS One 3, e2802.
DOI URL |
| [19] |
Hebert PDN, Cywinska A, Ball SL, deWaard JR (2003). Biological identifications through DNA barcodes.Proc Roy Soc B Biol Sci 270, 313-321.
DOI URL |
| [20] |
Hollingsworth PM (2008). DNA barcoding plants in biodiversity hot spots: progress and outstanding questions.Here- dity 101, 1-2.
DOI URL PMID |
| [21] | Kress WJ, Erickson DL (2007). A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS One 2, e508. |
| [22] |
Keller A, Schleicher T, Schultz J, Müller T, Dandekar T, Wolf M (2009). 5.8S-28S rRNA interaction and HMM-ba- sed ITS2 annotation.Gene 430, 50-57.
DOI URL PMID |
| [23] |
Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005). Use of DNA barcodes to identify flowering plants.Proc Natl Acad Sci USA 102, 8369-8374.
DOI URL PMID |
| [24] |
Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, Maurin O, Duthoit S, Barraclough TG, Savolainen V (2008). DNA barcoding the floras of bio- diversity hotspots.Proc Natl Acad Sci USA 105, 2923-2928.
DOI URL PMID |
| [25] |
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007). Clustal W and Clustal X version 2.0.Bioinformatics 23, 2947-2948.
DOI URL PMID |
| [26] |
Meyer CP, Paulay G (2005). DNA barcoding: error rates based on comprehensive sampling.PLoS Biol 3, e422.
DOI URL |
| [27] |
Newmaster SG, Fazekas AJ, Steeves RAD, Janovec J (2008). Testing candidate plant barcode regions in the Myris- ticaceae.Mol Ecol Res 8, 480-490.
DOI URL PMID |
| [28] |
Ross HA, Murugan S, Li WLS (2008). Testing the reliability of genetic methods of species identification via simulation.Syst Biol 57, 216-230.
DOI URL PMID |
| [29] |
Selig C, Wolf M, Müller T, Dandekar T, Schultz J (2008). The ITS2 Database II: homology modelling RNA structure for molecular systematics.Nucleic Acids Res 36, D377-D380.
DOI URL PMID |
| [30] |
Slabbinck B, Dawyndt P, Martens M, De Vos P, De Baets B (2008). Taxon gap: a visualization tool for intra- and inter- species variation among individual biomarkers.Bioinformatics 24, 866-867.
DOI URL PMID |
| [31] |
Song JY, Yao H, Li Y, Li XW, Lin YL, Liu C, Han JP, Xie CX, Chen SL (2009). Authentication of the family Polygonaceae in Chinese pharmacopoeia by DNA barcoding technique.J Ethnopharmacol 124, 434-439.
DOI URL PMID |
| [32] |
Vinitha MR, Kumar US, Aishwarya K, Sabu M, Thomas G (2014). Prospects for discriminating Zingiberaceae species in India using DNA barcodes.J Integr Plant Biol 56, 760-773.
DOI URL PMID |
| [33] |
Yao H, Song JY, Ma XY, Liu C, Li Y, Xu HX, Han JP, Duan LS, Chen SL (2009). Identification of Dendrobium species by a candidate DNA barcode sequence: the chloroplast psbA-trnH intergenic region. Planta Med 75, 667-669.
DOI URL PMID |
| [1] | Xiaoyan Luo, Qiang Li, Xiaolei Huang. DNA barcode reference dataset for flower-visiting insects in Daiyun Mountain National Nature Reserve [J]. Biodiv Sci, 2023, 31(8): 23236-. |
| [2] | Fan Wu, Shenyun Liu, Huqiang Jiang, Qian Wang, Kaiwei Chen, Hongliang Li. Pollination difference between Apis cerana cerana and Apis mellifera ligustica during the late autumn and winter [J]. Biodiv Sci, 2023, 31(5): 22528-. |
| [3] | Wang Lulu, Yang Zhi, Yang Yong. Plant Ultra-barcoding Using Herbariomics [J]. Chinese Bulletin of Botany, 2023, 58(5): 831-842. |
| [4] | Rong Sun, Yulu Yang, Yajun Li, Hui Zhang, Xukai Li. Genome-wide Identification and Analysis of PLATZ Transcription Factor Gene Family in Foxtail Millet [J]. Chinese Bulletin of Botany, 2023, 58(4): 548-559. |
| [5] | Jiajia Pu, Pingjun Yang, Yang Dai, Kexin Tao, Lei Gao, Yuzhou Du, Jun Cao, Xiaoping Yu, Qianqian Yang. Species identification and population genetic structure of non-native apple snails (Ampullariidea: Pomacea) in the lower reaches of the Yangtze River [J]. Biodiv Sci, 2023, 31(3): 22346-. |
| [6] | 春惠 林 huiyi gu qinliang ye zhijian zhang Zhong Zhiming 绮斐 易. Population structure and dynamic characteristics of endangered Camellia granthamiana Sealy [J]. Chin J Plant Ecol, 2023, 47(12): 1684-1692. |
| [7] | Jie Tao, Benqiang Li, Jinghua Cheng, Ying Shi, Peihong Liu, Guixia He, Weijie Xu, Huili Liu. Viral metagenome analysis of the viral community composition of the porcine diarrhea feaces [J]. Biodiv Sci, 2023, 31(11): 23170-. |
| [8] | Keyi Wu, Wenda Ruan, Difeng Zhou, Qingchen Chen, Chengyun Zhang, Xinyuan Pan, Shang Yu, Yang Liu, Rongbo Xiao. Syllable clustering analysis-based passive acoustic monitoring technology and its application in bird monitoring [J]. Biodiv Sci, 2023, 31(1): 22370-. |
| [9] | Ke Deng, Jianguo Cui. Application and prospect of network analysis in the studies of animal vocal communication and bioacoustics [J]. Biodiv Sci, 2023, 31(1): 22318-. |
| [10] | Yuzhuo Wang, Yuanxiang Lin, Yapeng Xue, Zhengyong Duan, Xiaodan Wang, Ling Chen, Xiaoning Cao, Ruiyun Wang, Zhijun Qiao. Construction of Molecular ID Card of Core Germplasm of Hog Millet (Panicum miliaceum) in Shanxi [J]. Chinese Bulletin of Botany, 2023, 58(1): 159-168. |
| [11] | Yajing Song, Jinwen Ou, Guwen Zhang, Zhijuan Feng, Yuanpeng Bu, Bin Wang, Yaming Gong, Jianqiang Xu, Na Liu. Pathogen Identification of Pea Crown Rot and Its Sensitivity to Fungicides [J]. Chinese Bulletin of Botany, 2023, 58(1): 132-139. |
| [12] | Xiaofeng Niu, Xiaomei Wang, Yan Zhang, Zhipeng Zhao, Enyuan Fan. Integration and application of sturgeon identification methods [J]. Biodiv Sci, 2022, 30(6): 22034-. |
| [13] | Cong Xu, Feiyu Zhang, Daoyuan Yu, Xin Sun, Feng Zhang. Performance evaluation of molecular taxonomy assignment tools for soil invertebrates [J]. Biodiv Sci, 2022, 30(12): 22252-. |
| [14] | Yixin Sun, Yingbin Li, Yuhui Li, Bing Li, Xiaofang Du, Qi Li. Application of high-throughput sequencing technique in the study of nematode diversity [J]. Biodiv Sci, 2022, 30(12): 22266-. |
| [15] | WANG Chun-Cheng, ZHANG Yun-Ling, MA Song-Mei, HUANG Gang, ZHANG Dan, YAN Han. Phylogeny and species differentiation of four wild almond species of subgen. Amygdalus in China [J]. Chin J Plant Ecol, 2021, 45(9): 987-995. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||