

Chinese Bulletin of Botany ›› 2017, Vol. 52 ›› Issue (6): 689-698.DOI: 10.11983/CBB17132
• INVITED REVIEWS • Previous Articles Next Articles
Liu Jingyan, Shi Yiting, Yang Shuhua*( )
)
Received:2017-07-13
Accepted:2017-09-12
Online:2017-11-01
Published:2018-12-10
Contact:
Yang Shuhua
Liu Jingyan, Shi Yiting, Yang Shuhua. CBF: A Key Factor Balancing Plant Cold Stress Responses and Growth[J]. Chinese Bulletin of Botany, 2017, 52(6): 689-698.
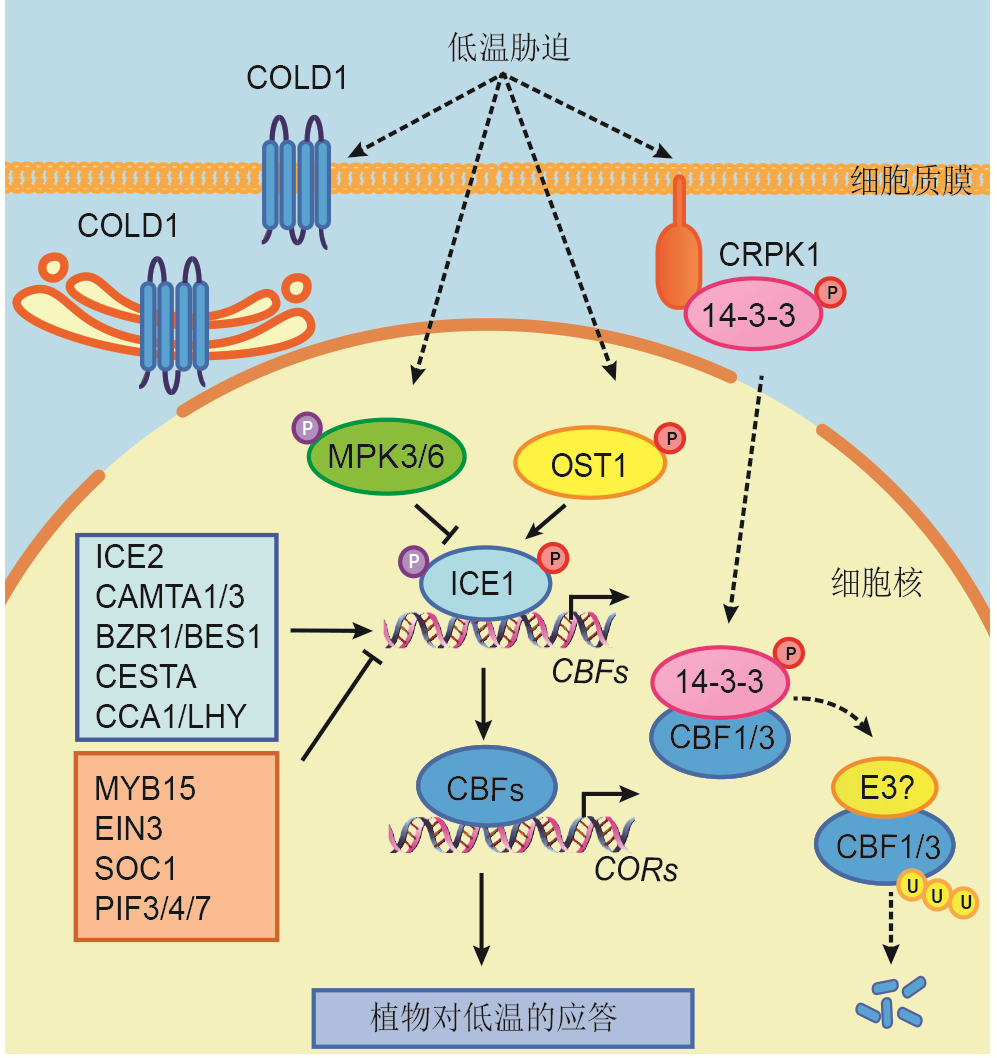
Figure 1 CBF-dependent cold signaling pathwayThe solid lines indicate direct regulation, and dotted lines indicate indirect regulation. Positive regulation is indicated by arrow heads and negative regulation is indicated by T-shaped lines.
| [1] |
Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P (2008). The cold-inducible CBF1 factor: dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism.Plant Cell 20, 2117-2129.
DOI URL |
| [2] | Agarwal M, Hao YJ, Kapoor A, Dong CH, Fujii H, Zheng XW, Zhu JK (2006). A R2R3 type MYB transcription factor is involved in the cold regulation ofCBF genes and in acquired freezing tolerance. J Biol Chem 281, 37636-37645. |
| [3] | Baker SS, Wilhelm KS, Thomashow MF (1994). The 5'- region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol 24, 701-713. |
| [4] |
Bieniawska Z, Espinoza C, Schlereth A, Sulpice R, Hincha DK, Hannah MA (2008). Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome.Plant Physiol 147, 263-279.
DOI URL PMID |
| [5] | Blázquez MA, Ahn JH, Weigel D (2003). A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet 33, 168-171. |
| [6] |
Canella D, Gilmour SJ, Kuhn LA, Thomashow MF (2010). DNA binding by the Arabidopsis CBF1 transcription factor requires the PKKP/RAGRxKFxETRHP signature sequen- ce.Biochim Biophys Acta 1799, 454-462.
DOI URL PMID |
| [7] |
Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong XH, Agarwal M, Zhu JK (2003). ICE1: a regulator of cold- induced transcriptome and freezing tolerance in Arabidopsis.Genes Dev 17, 1043-1054.
DOI URL PMID |
| [8] |
Deng CY, Ye HY, Fan M, Pu TL, Yan JB (2017). The rice transcription factors OsICE confer enhanced cold tolerance in transgenic Arabidopsis.Plant Signal Behav 12, e1316442.
DOI URL PMID |
| [9] |
Ding YL, Li H, Zhang XY, Xie Q, Gong ZZ, Yang SH (2015). OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis.Dev Cell 32, 278-289.
DOI URL PMID |
| [10] |
Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance.Plant Cell 21, 972-984.
DOI URL |
| [11] |
Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1.Proc Natl Acad Sci USA 103, 8281-8286.
DOI URL |
| [12] |
Dong MA, Farré EM, Thomashow MF (2011). Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis.Proc Natl Acad Sci USA 108, 7241-7246.
DOI URL PMID |
| [13] |
Eremina M, Unterholzner SJ, Rathnayake AI, Castellanos M, Khan M, Kugler KG, May ST, Mayer KFX, Rozhon W, Poppenberger B (2016). Brassinosteroids participate in the control of basal and acquired freezing tolerance of plants.Proc Natl Acad Sci USA 113, E5982-E5991.
DOI URL PMID |
| [14] |
Espinoza C, Bieniawska Z, Hincha DK, Hannah MA (2008). Interactions between the circadian clock and cold- response in Arabidopsis.Plant Signal Behav 3, 593-594.
DOI URL PMID |
| [15] | Franklin KA, Whitelam GC (2007). Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat Genet 39, 1410-1413. |
| [16] | Fursova OV, Pogorelko GV, Tarasov VA (2009). Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Ge- ne 429, 98-103. |
| [17] |
Gilmour SJ, Fowler SG, Thomashow MF (2004). Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities.Plant Mol Biol 54, 767-781.
DOI URL |
| [18] | Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR ge- ne expression. Plant J 16, 433-442. |
| [19] |
Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock.Science 290, 2110-2113.
DOI URL PMID |
| [20] |
He JX, Gendron JM, Sun Y, Gampala SSL, Gendron N, Sun CQ, Wang ZY (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses.Science 307, 1634-1638.
DOI URL PMID |
| [21] |
Hong JH, Savina M, Du J, Devendran A, Ramakanth KK, Tian X, Sim WS, Mironova VV, Xu J (2017). A sacrifice-for-survival mechanism protects root stem cell niche from chilling stress.Cell 170, 102-113.
DOI URL PMID |
| [22] |
Hu Y, Jiang LQ, Wang F, Yu DQ (2013). Jasmonate regulates the INDUCER OF CBF EXPRESSION-C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 cascade and freezing tolerance in Arabidopsis.Plant Cell 25, 2907-2924.
DOI URL PMID |
| [23] | Jaglo KR, Kleff S, Amundsen KL, Zhang X, Haake V, Zhang JZ, Deits T, Thomashow MF (2001). Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol 127, 910-917. |
| [24] |
Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tole- rance. Science 280, 104-106.
DOI URL PMID |
| [25] |
Jia YX, Ding YL, Shi YT, Zhang XY, Gong ZZ, Yang SH (2016). The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol 212, 345-353.
DOI URL PMID |
| [26] |
Jiang BC, Shi YT, Zhang XY, Xin XY, Qi LJ, Guo HW, Li JG, Yang SH (2017). PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc Natl Acad Sci USA 114, E6695-E6702.
DOI URL PMID |
| [27] |
Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao MJ, Khattak AK, Box MS, Charoensawan V, Cortijo S, Kumar M, Grant A, Locke JCW, Sch?fer E, Jaeger KE, Wigge PA (2016). Phytochromes function as thermosensors in Arabidopsis.Science 354, 886-889.
DOI URL PMID |
| [28] |
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor.Nat Biotechnol 17, 287-291.
DOI URL PMID |
| [29] | Kasuga M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K (2004). A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol 45, 346-350. |
| [30] |
Kidokoro S, Yoneda K, Takasaki H, Takahashi F, Shinozaki K, Yamaguchi-Shinozaki K (2017). Different cold- signaling pathways function in the responses to rapid and gradual decreases in temperature.Plant Cell 29, 760-774.
DOI URL PMID |
| [31] |
Kim SH, Kim HS, Bahk S, An J, Yoo Y, Kim JY, Chung WS (2017). Phosphorylation of the transcriptional repressor MYB15 by mitogen-activated protein kinase 6 is required for freezing tolerance in Arabidopsis.Nucleic Acids Res 45, 6613-6627.
DOI URL PMID |
| [32] |
Kim Y, Park S, Gilmour SJ, Thomashow MF (2013). Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis.Plant J 75, 364-376.
DOI URL PMID |
| [33] |
Lee BH, Henderson DA, Zhu JK (2005). The Arabidopsis cold-responsive transcriptome and its regulation by ICE1.Plant Cell 17, 3155-3175.
DOI URL PMID |
| [34] |
Lee CM, Thomashow MF (2012). Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance inArabidopsis thaliana. Proc Natl Acad Sci USA 109, 15054-15059.
DOI URL PMID |
| [35] |
Legris M, Klose C, Burgie ES, Rojas CCR, Neme M, Hiltbrunner A, Wigge PA, Sch?fer E, Vierstra RD, Casal JJ (2016). Phytochrome B integrates light and temperature signals in Arabidopsis.Science 354, 897-900.
DOI URL PMID |
| [36] |
Leivar P, Monte E (2014). PIFs: systems integrators in plant development.Plant Cell 26, 56-78.
DOI URL |
| [37] |
Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness.Curr Biol 18, 1815-1823.
DOI URL PMID |
| [38] | Li H, Ding YL, Shi YT, Zhang XY, Zhang SQ, Gong ZZ, Yang SH (2017a). MPK3- and MPK6-mediated ICE1 pho- sphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis.Dev Cell. doi:10.1016/ j.devcel.2017.09.025. |
| [39] |
Li H, Ye KY, Shi YT, Cheng JK, Zhang XY, Yang SH (2017b). BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabi- dopsis.Mol Plant 10, 545-559.
DOI URL PMID |
| [40] |
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive ge- ne expression, respectively, in Arabidopsis.Plant Cell 10, 1391-1406.
DOI URL PMID |
| [41] |
Liu ZY, Jia YX, Ding YL, Shi YT, Li Z, Guo Y, Gong ZZ, Yang SH (2017). Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response.Mol Cell 66, 117-128.
DOI URL PMID |
| [42] | Lu X, Yang L, Yu MY, Lai JB, Wang C, McNeil D, Zhou MX, Yang CW (2017). A novel Zea mays ssp. mexicana L. MYC-type ICE-like transcription factor gene ZmmICE1, en- hances freezing tolerance in transgenic Arabidopsis tha- liana. Plant Physiol Biochem 113, 78-88. |
| [43] | Ma Y, Dai XY, Xu YY, Luo W, Zheng XM, Zeng DL, Pan YJ, Lin XL, Liu HH, Zhang DJ, Xiao J, Guo XY, Xu SJ, Niu YD, Jin JB, Zhang H, Xu X, Li LG, Wang W, Qian Q, Ge S, Chong K (2015). COLD1 confers chilling tolerance in rice. Cell 160, 1209-1221. |
| [44] |
Medina J, Bargues M, Terol J, Perez-Alonso M, Salinas J (1999). The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins who- se expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol 119, 463-470.
DOI URL PMID |
| [45] |
Medina J, Catalá R, Salinas J (2011). The CBFs: three Arabidopsis transcription factors to cold acclimate.Plant Sci 180, 3-11.
DOI URL PMID |
| [46] | Meshi T, Iwabuchi M (1995). Plant transcription factors.Plant Cell Physiol 36, 1405-1420. |
| [47] |
Mikkelsen MD, Thomashow MF (2009). A role for circadian evening elements in cold-regulated gene expression in Arabidopsis.Plant J 60, 328-339.
DOI URL PMID |
| [48] |
Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ash- worth EN, Bressan RA, Yun DJ, Hasegawa PM (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DRE- B1A expression and freezing tolerance in Arabidopsis. Plant Cell 19, 1403-1414.
DOI URL PMID |
| [49] |
Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, Sakakibara H, Mizuno T (2012). Trans- criptional repressor PRR5 directly regulates clock-output pathways.Proc Natl Acad Sci USA 109, 17123-17128.
DOI URL |
| [50] |
Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, Saito K, Sakakibara H, Mizuno T (2009). Transcript profiling of an Arabidopsis PSEUDO RESP- ONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol 50, 447-462.
DOI URL PMID |
| [51] |
Ni M, Tepperman JM, Quail PH (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein.Cell 95, 657-667.
DOI URL PMID |
| [52] |
Ni WM, Xu SL, González-Grandío E, Chalkley RJ, Huhmer AFR, Burlingame AL, Wang ZY, Quail PH (2017). PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3.Nat Commun 8, 15236.
DOI URL PMID |
| [53] |
Ni WM, Xu SL, Tepperman JM, Stanley DJ, Maltby DA, Gross JD, Burlingame AL, Wang ZY, Quail PH (2014). A mutually assured destruction mechanism attenuates light signaling in Arabidopsis.Science 344, 1160-1164.
DOI URL PMID |
| [54] | Nosenko T, B?ndel KB, Kumpfmüller G, Stephan W (2016). Adaptation to low temperatures in the wild tomato speciesSolanum chilense. Mol Ecol 25, 2853-2869. |
| [55] |
Novillo F, Alonso JM, Ecker JR, Salinas J (2004). CBF2/ DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc Natl Acad Sci USA 101, 3985-3990.
DOI URL PMID |
| [56] |
Novillo F, Medina J, Salinas J (2007). Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon.Proc Natl Acad Sci USA 104, 21002-21007.
DOI URL PMID |
| [57] |
Ohme-Takagi M, Shinshi H (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element.Plant Cell 7, 173-182.
DOI URL PMID |
| [58] |
Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis.Proc Natl Acad Sci USA 94, 7076-7081.
DOI URL |
| [59] |
Park S, Lee CM, Doherty CJ, Gilmour SJ, Kim Y, Thom- ashow MF (2015). Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network.Plant J 82, 193-207.
DOI URL PMID |
| [60] |
Qin F, Sakuma Y, Li J, Liu Q, Li YQ, Shinozaki K, Yamaguchi-Shinozaki K (2004). Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L. Plant Cell Physiol 45, 1042-1052.
DOI URL PMID |
| [61] |
Riechmann JL, Meyerowitz EM (1998). The AP2/EREBP family of plant transcription factors.Biol Chem 379, 633-646.
DOI URL PMID |
| [62] |
Seo E, Lee H, Jeon J, Park H, Kim J, Noh YS, Lee I (2009). Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell 21, 3185-3197.
DOI URL PMID |
| [63] |
Seo PJ, Park MJ, Lim MH, Kim SG, Lee M, Baldwin IT, Park CM (2012). A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock re- gulation of temperature responses in Arabidopsis.Plant Cell 24, 2427-2442.
DOI URL |
| [64] |
Shi YT, Tian SW, Hou LY, Huang XZ, Zhang XY, Guo HW, Yang SH (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24, 2578-2595.
DOI URL PMID |
| [65] |
Shimura Y, Shiraiwa Y, Suzuki I (2012). Characterization of the subdomains in the N-terminal region of histidine kinase Hik33 in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol 53, 1255-1266.
DOI URL PMID |
| [66] | Stockinger EJ, Gilmour SJ, Thomashow MF (1997). Ara- bidopsis thaliana CBF1 encodes an AP2 domaincon- taining transcriptional activator that binds to the C-repeat/ DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94, 1035-1040. |
| [67] |
Yin YH, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J (2005). A new class of transcription factors mediates br- assinosteroid-regulated gene expression in Arabidopsis.Cell 120, 249-259.
DOI URL PMID |
| [68] | Zabulon G, Richaud C, Guidi-Rontani C, Thomas JC (2007). NblA gene expression in Synechocystis PCC 6803 strains lacking DspA (Hik33) and a NblR-like protein. Curr Microbiol 54, 36-41. |
| [69] |
Zhao CZ, Wang PC, Si T, Hsu CC, Wang L, Zayed O, Yu ZP, Zhu YF, Dong J, Tao WA, Zhu JK (2017). MAP kinase cascades regulate the cold response by modulating ICE1 protein stability.Dev Cell. doi: 10.1016/j.devcel. 2017.09.024
DOI URL PMID |
| [70] |
Zhao CZ, Zhang ZJ, Xie SJ, Si T, Li YY, Zhu JK (2016). Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis.Plant Phy- siol 171, 2744-2759.
DOI URL PMID |
| [1] | Xinhai Zeng, Rui Chen, Yu Shi, Chaoyue Gai, Kai Fan, Zhaowei Li. Research Advances in Biological Functions of Plant SPL Transcription Factors [J]. Chinese Bulletin of Botany, 2023, 58(6): 982-997. |
| [2] | Yanan Xu, Jiarong Yan, Xin Sun, Xiaomei Wang, Yufeng Liu, Zhouping Sun, Mingfang Qi, Tianlai Li, Feng Wang. Red and Far-red Light Regulation of Plant Growth, Development, and Abiotic Stress Responses [J]. Chinese Bulletin of Botany, 2023, 58(4): 622-637. |
| [3] | Jia Zhang, Qidong Li, Cui Li, Qinghai Wang, Xincun Hou, Chunqiao Zhao, Shuhe Li, Qiang Guo. Research Progress on MATE Transporters in Plants [J]. Chinese Bulletin of Botany, 2023, 58(3): 461-474. |
| [4] | Qi Wang, Yunzhe Wu, Xueying Liu, Lili Sun, Hong Liao, Xiangdong Fu. The Rice Receptor-like Kinases Function as Key Regulators of Plant Development and Adaptation to the Environment [J]. Chinese Bulletin of Botany, 2023, 58(2): 199-213. |
| [5] | Deshuai Liu, Lei Yao, Weirong Xu, Mei Feng, Wenkong Yao. Research Progress of Melatonin in Plant Stress Resistance [J]. Chinese Bulletin of Botany, 2022, 57(1): 111-126. |
| [6] | Jianru Yue, Yunjian He, Tianqi Qiu, Nannan Guo, Xueping Han, Xianling Wang. Research Advances in the Molecular Mechanisms of Plant Microtubules in Regulating Hypocotyl Elongation [J]. Chinese Bulletin of Botany, 2021, 56(3): 363-371. |
| [7] | Qilu Yu, Jiangzhe Zhao, Xiaoxian Zhu, Kewei Zhang. Regulation of Rice Growth by Root-secreted Phytohormones [J]. Chinese Bulletin of Botany, 2021, 56(2): 175-182. |
| [8] | Xibao Li, Minyi Lai, Shan Liang, Xiaojing Wang, Caiji Gao, Chao Yang. Function and Transcriptional Regulation of Autophagy-related Genes in Plants [J]. Chinese Bulletin of Botany, 2021, 56(2): 201-217. |
| [9] | Jindong Wang,Yu Zhou,Jiawen Yu,Xiaolei Fan,Changquan Zhang,Qianfeng Li,Qiaoquan Liu. Advances in the Regulation of Plant Growth and Development and Stress Response by miR172-AP2 Module [J]. Chinese Bulletin of Botany, 2020, 55(2): 205-215. |
| [10] | Menglong Wang,Xiaoqun Peng,Zhufeng Chen,Xiaoyan Tang. Research Advances on Lectin Receptor-like Kinases in Plants [J]. Chinese Bulletin of Botany, 2020, 55(1): 96-105. |
| [11] | Qianqian Guo,Wenbin Zhou. Advances in the Mechanism Underlying Plant Response to Stress Combination [J]. Chinese Bulletin of Botany, 2019, 54(5): 662-673. |
| [12] | Du Kangxi, Shen Wenhui, Dong Aiwu. Advances in Epigenetic Regulation of Abiotic Stress Response in Plants [J]. Chinese Bulletin of Botany, 2018, 53(5): 581-593. |
| [13] | Wei Hua, Wang Yan, Liu Baohui, Wang Lei. Deciphering the Underlying Mechanism of the Plant Circadian System and Its Regulation on Plant Growth and Development [J]. Chinese Bulletin of Botany, 2018, 53(4): 456-467. |
| [14] | Yanqun Li, Hong Wu. The Research Progress of the Correlation Between Growth Development and Dynamic Accumulation of the Effective Components in Medicinal Plants [J]. Chinese Bulletin of Botany, 2018, 53(3): 293-304. |
| [15] | Han Danlu, Lai Jianbin, Yang Chengwei. Research Advances in Functions of SUMO E3 Ligases in Plant Growth and Development [J]. Chinese Bulletin of Botany, 2018, 53(2): 175-184. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||