

Chinese Bulletin of Botany ›› 2018, Vol. 53 ›› Issue (1): 59-71.DOI: 10.11983/CBB16257
• EXPERIMENTAL COMMUNICATIONS • Previous Articles Next Articles
Guodong Wu, Yu Xiu, Huafang Wang*( )
)
Received:2016-12-23
Accepted:2017-05-04
Online:2018-01-01
Published:2018-08-10
Contact:
Huafang Wang
Guodong Wu, Yu Xiu, Huafang Wang. Breeding of MtDREB2A Transgenic Soybean by an Optimized Cotyledonary-Node Method[J]. Chinese Bulletin of Botany, 2018, 53(1): 59-71.
| Primer name | Primer sequence (5′-3′) | Tm (°C) | Product length (bp) |
|---|---|---|---|
| rd29A-F1 | GGCTTTACACTTTATGCTTCC | 49.2 | 859 |
| rd29A-F2 | TTGTTAGGCTCCCTCATTTC | ||
| rd29A-T1 | CAGTTTGAAAGAAAAGGGAA | 46.7 | 71 |
| rd29A-T2 | GCTTTTTGGAACTCATGTCG | ||
| MtDREB2A-F1 | CATGCCATGGTGGAAATTGAAAGATGGGTGCT | 53.5 | 971 |
| MtDREB2A-F2 | GGGTGACCGGATTATTATCTAGTTGCCCAAACG | ||
| MtDREB2A-T1 | ACTTTTCCGACGGCTCAA | 44.0 | 472 |
| MtDREB2A-T2 | GTCATTACACACACCCTCTC |
Table 1 The primers used in construction of plant express vector
| Primer name | Primer sequence (5′-3′) | Tm (°C) | Product length (bp) |
|---|---|---|---|
| rd29A-F1 | GGCTTTACACTTTATGCTTCC | 49.2 | 859 |
| rd29A-F2 | TTGTTAGGCTCCCTCATTTC | ||
| rd29A-T1 | CAGTTTGAAAGAAAAGGGAA | 46.7 | 71 |
| rd29A-T2 | GCTTTTTGGAACTCATGTCG | ||
| MtDREB2A-F1 | CATGCCATGGTGGAAATTGAAAGATGGGTGCT | 53.5 | 971 |
| MtDREB2A-F2 | GGGTGACCGGATTATTATCTAGTTGCCCAAACG | ||
| MtDREB2A-T1 | ACTTTTCCGACGGCTCAA | 44.0 | 472 |
| MtDREB2A-T2 | GTCATTACACACACCCTCTC |
| Germination medium | Re-suspension medium | Co-cultivation medium | Shoot induction medium | Shoot elongation medium | Rooting medium | |
|---|---|---|---|---|---|---|
| MS salts | 1/2 × | 1/2 × | - | - | - | - |
| MS iron stock | 1/2 × | 1/2 × | 1 × | 1 × | 1 × | 1 × |
| MS vitamins | 1/2 × | 1/2 × | - | - | - | - |
| B5 salts | - | - | 1 × | 1 × | 1 × | 1/2 × |
| B5 vitamins | - | - | 1 × | 1 × | 1 × | - |
| Sucrose (g·L-1) | 15 | - | 30 | 30 | 30 | 30 |
| Glucose (g·L-1) | - | 10 | - | - | - | - |
| Agar (g·L-1) | 8 | - | 5 | 8 | 8 | 8 |
| pH | 5.8 | 5.8 | 5.5 | 5.5 | 5.5 | 5.6 |
| 6-BA (mg·L-1) | - | - | 1.7 | 1.7 | 1.7 | - |
| GA (mg·L-1) | - | - | - | - | 1.0 | - |
| IBA (mg·L-1) | - | - | - | - | - | 1.0 |
| MES (g·L-1) | - | - | 0.6 | 0.6 | 0.6 | 0.6 |
| L-cys (mg·L-1) | - | - | 182.5 | 182.5 | 182.5 | - |
| Na2S2O3 (mg·L-1) | - | - | 250 | 250 | 250 | - |
| DTT (mg·L-1) | - | - | 154.3 | - | - | - |
| AS (mg·L-1) | - | 39.2 | 39.2 | - | - | - |
| Cef (mg·L-1) | - | - | - | 100-400 | 100-400 | - |
| Cb (mg·L-1) | - | - | - | 100-400 | 100-400 | - |
| PPT (mg·L-1) | - | - | - | 0, 2-5 | - | - |
Table 2 List of components used in media preparation for transformation method
| Germination medium | Re-suspension medium | Co-cultivation medium | Shoot induction medium | Shoot elongation medium | Rooting medium | |
|---|---|---|---|---|---|---|
| MS salts | 1/2 × | 1/2 × | - | - | - | - |
| MS iron stock | 1/2 × | 1/2 × | 1 × | 1 × | 1 × | 1 × |
| MS vitamins | 1/2 × | 1/2 × | - | - | - | - |
| B5 salts | - | - | 1 × | 1 × | 1 × | 1/2 × |
| B5 vitamins | - | - | 1 × | 1 × | 1 × | - |
| Sucrose (g·L-1) | 15 | - | 30 | 30 | 30 | 30 |
| Glucose (g·L-1) | - | 10 | - | - | - | - |
| Agar (g·L-1) | 8 | - | 5 | 8 | 8 | 8 |
| pH | 5.8 | 5.8 | 5.5 | 5.5 | 5.5 | 5.6 |
| 6-BA (mg·L-1) | - | - | 1.7 | 1.7 | 1.7 | - |
| GA (mg·L-1) | - | - | - | - | 1.0 | - |
| IBA (mg·L-1) | - | - | - | - | - | 1.0 |
| MES (g·L-1) | - | - | 0.6 | 0.6 | 0.6 | 0.6 |
| L-cys (mg·L-1) | - | - | 182.5 | 182.5 | 182.5 | - |
| Na2S2O3 (mg·L-1) | - | - | 250 | 250 | 250 | - |
| DTT (mg·L-1) | - | - | 154.3 | - | - | - |
| AS (mg·L-1) | - | 39.2 | 39.2 | - | - | - |
| Cef (mg·L-1) | - | - | - | 100-400 | 100-400 | - |
| Cb (mg·L-1) | - | - | - | 100-400 | 100-400 | - |
| PPT (mg·L-1) | - | - | - | 0, 2-5 | - | - |
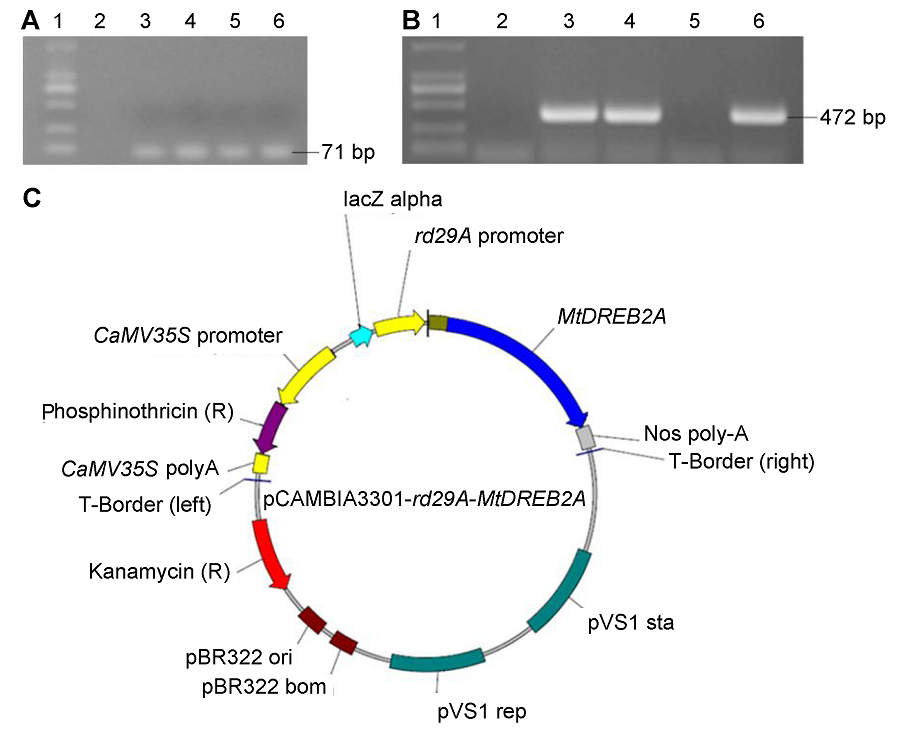
Figure 1 Construction of plant express vector pCAMBIA3301-rd29A-MtDREB2A(A) PCR confirmation of rd29A promoter (1: Marker; 2: pCAMBIA3301-35S-GUS; 3-6: PCR product of rd29A); (B) PCR confirmation of MtDREB2A genes (1: Marker; 2: pCAMBIA3301-35S-GUS; 3-6: PCR product of MtDREB2A); (C) Structure of pCAMBIA3301-rd29A-MtDREB2A
| Treatment | NaClO concentration (%) | Time (min) | Sterilization rate (%) | Contamination rate (%) | Death rate (%) |
|---|---|---|---|---|---|
| 1 | 0.10 | 3 | 81.33±3.06 e | 16.00±2.00 b | 2.67±1.16 bcd |
| 2 | 0.10 | 5 | 72.67±1.16 f | 24.00±2.00 a | 3.33±1.16 bc |
| 3 | 0.10 | 10 | 88.67±1.16 bc | 10.67±1.16 cd | 0.67±1.16 d |
| 4 | 0.25 | 3 | 86.67±1.16 cd | 10.00±2.00 cde | 3.33±1.16 bc |
| 5 | 0.25 | 5 | 70.67±2.31 f | 23.33±4.16 a | 6.00±2.00 a |
| 6 | 0.25 | 10 | 90.67±1.16 b | 8.00±2.00 de | 1.33±1.16 cd |
| 7 | 0.50 | 3 | 83.33±3.06 de | 13.33±3.06 bc | 3.33±1.16 bc |
| 8 | 0.50 | 5 | 89.33±2.31 bc | 6.00±2.00 ef | 4.67±1.16 ab |
| 9 | 0.50 | 10 | 98.67±1.16 a | 0.67±1.16 g | 0.67±1.16 d |
Table 3 Effect of NaClO method on seed sterilization of Glycine max cv. ‘Dongnong 50’ (means±SD)
| Treatment | NaClO concentration (%) | Time (min) | Sterilization rate (%) | Contamination rate (%) | Death rate (%) |
|---|---|---|---|---|---|
| 1 | 0.10 | 3 | 81.33±3.06 e | 16.00±2.00 b | 2.67±1.16 bcd |
| 2 | 0.10 | 5 | 72.67±1.16 f | 24.00±2.00 a | 3.33±1.16 bc |
| 3 | 0.10 | 10 | 88.67±1.16 bc | 10.67±1.16 cd | 0.67±1.16 d |
| 4 | 0.25 | 3 | 86.67±1.16 cd | 10.00±2.00 cde | 3.33±1.16 bc |
| 5 | 0.25 | 5 | 70.67±2.31 f | 23.33±4.16 a | 6.00±2.00 a |
| 6 | 0.25 | 10 | 90.67±1.16 b | 8.00±2.00 de | 1.33±1.16 cd |
| 7 | 0.50 | 3 | 83.33±3.06 de | 13.33±3.06 bc | 3.33±1.16 bc |
| 8 | 0.50 | 5 | 89.33±2.31 bc | 6.00±2.00 ef | 4.67±1.16 ab |
| 9 | 0.50 | 10 | 98.67±1.16 a | 0.67±1.16 g | 0.67±1.16 d |
| Treatment | Time (h) | Sterilization rate (%) | Contamination rate (%) | Death rate (%) |
|---|---|---|---|---|
| 1 | 1 | 82.00±5.29 b | 13.33±5.78 a | 4.66±1.16 b |
| 2 | 2 | 82.67±6.11 b | 12.00±6.00 a | 5.33±3.06 b |
| 3 | 4 | 92.00±3.46 a | 2.67±2.31 b | 5.33±3.06 b |
| 4 | 6 | 93.33±1.16 a | 2.67±2.31 b | 4.00±2.00 b |
| 5 | 8 | 88.00±3.46 ab | 1.33±2.31 b | 10.67±1.16 a |
Table 4 Effect of Cl2 method on seed sterilization of Glycine max cv. ‘Dongnong 50’
| Treatment | Time (h) | Sterilization rate (%) | Contamination rate (%) | Death rate (%) |
|---|---|---|---|---|
| 1 | 1 | 82.00±5.29 b | 13.33±5.78 a | 4.66±1.16 b |
| 2 | 2 | 82.67±6.11 b | 12.00±6.00 a | 5.33±3.06 b |
| 3 | 4 | 92.00±3.46 a | 2.67±2.31 b | 5.33±3.06 b |
| 4 | 6 | 93.33±1.16 a | 2.67±2.31 b | 4.00±2.00 b |
| 5 | 8 | 88.00±3.46 ab | 1.33±2.31 b | 10.67±1.16 a |
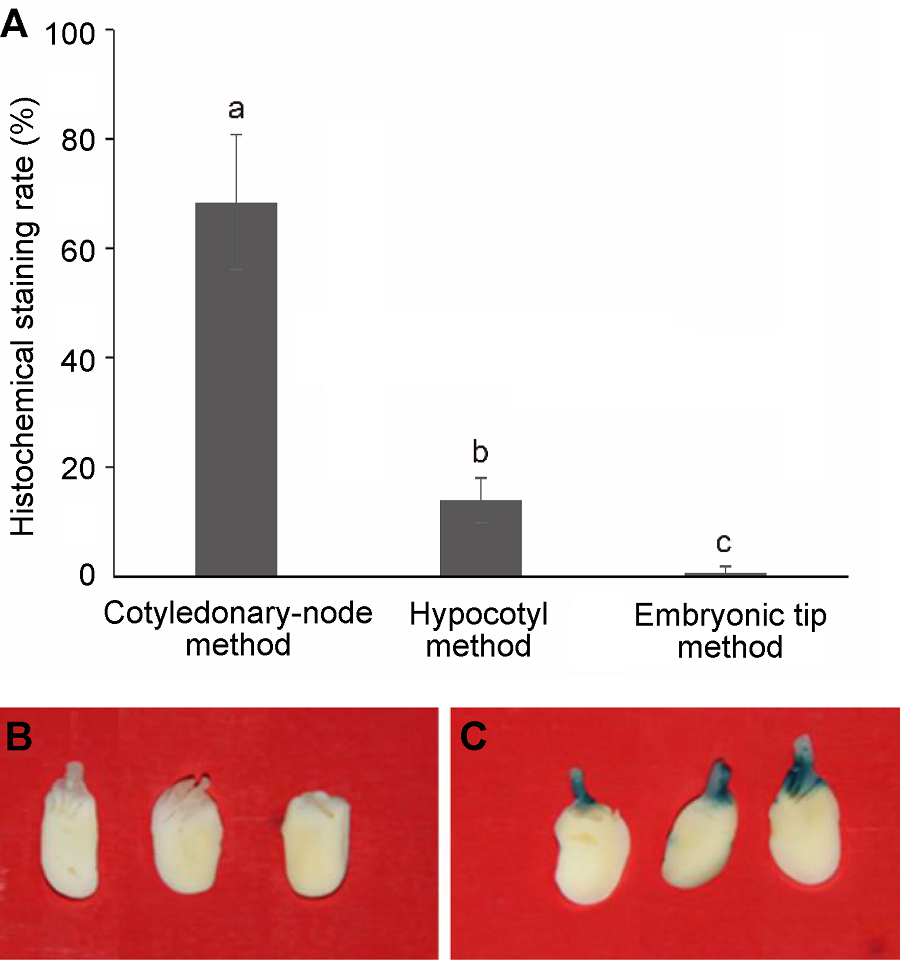
Figure 2 GUS gene histochemical staining(A) The effect of transgenic methods on histochemical GUS staining rate, different lowercase letters indicate significant differences at 0.05 level according to Duncan’s test; (B), (C) The GUS gene histochemical staining of cotyledonary node after co-cultivation ((B) Negative control; (C) Positive result of histochemical stain)
| Treatment | Factor | Histochemical staining rate of cotyledonary-node (%) | |||
|---|---|---|---|---|---|
| Germination time (d) (A) | Culture temperature (°C) (B) | OD600 value (C) | Co-cultivation time (d) (D) | ||
| 1 | 3 | 25 | 0.3 | 2 | 8.00±2.00 g |
| 2 | 3 | 26 | 0.5 | 3 | 31.05±10.46 def |
| 3 | 3 | 27 | 0.7 | 4 | 38.27±5.46 cde |
| 4 | 3 | 28 | 0.9 | 5 | 46.30±17.65 bcd |
| 5 | 4 | 25 | 0.5 | 4 | 39.70±19.34 cde |
| 6 | 4 | 26 | 0.3 | 5 | 59.33±3.06 ab |
| 7 | 4 | 27 | 0.9 | 2 | 4.00±2.00 g |
| 8 | 4 | 28 | 0.7 | 3 | 21.33±6.43 efg |
| 9 | 5 | 25 | 0.7 | 5 | 68.33±12.42 a |
| 10 | 5 | 26 | 0.9 | 4 | 51.06±7.65 abc |
| 11 | 5 | 27 | 0.3 | 3 | 30.56±16.17 def |
| 12 | 5 | 28 | 0.5 | 2 | 4.00±5.29 g |
| 13 | 6 | 25 | 0.9 | 3 | 40.95±11.61 bcd |
| 14 | 6 | 26 | 0.7 | 2 | 8.52±1.70 g |
| 15 | 6 | 27 | 0.5 | 5 | 20.36±9.30 efg |
| 16 | 6 | 28 | 0.3 | 4 | 12.67±10.26 fg |
| K1 | 30.91 | 39.25 | 27.64 | 6.13 | |
| K2 | 31.09 | 37.49 | 23.78 | 30.97 | |
| K3 | 38.49 | 23.30 | 34.12 | 35.43 | |
| K4 | 20.62 | 21.07 | 35.58 | 48.58 | |
| R | 17.87 | 18.18 | 11.80 | 42.45 | |
Table 5 The results of orthogonal experiment for Agrobacterium-mediated transformation of Glycine max cv. ‘Dongnong 50’
| Treatment | Factor | Histochemical staining rate of cotyledonary-node (%) | |||
|---|---|---|---|---|---|
| Germination time (d) (A) | Culture temperature (°C) (B) | OD600 value (C) | Co-cultivation time (d) (D) | ||
| 1 | 3 | 25 | 0.3 | 2 | 8.00±2.00 g |
| 2 | 3 | 26 | 0.5 | 3 | 31.05±10.46 def |
| 3 | 3 | 27 | 0.7 | 4 | 38.27±5.46 cde |
| 4 | 3 | 28 | 0.9 | 5 | 46.30±17.65 bcd |
| 5 | 4 | 25 | 0.5 | 4 | 39.70±19.34 cde |
| 6 | 4 | 26 | 0.3 | 5 | 59.33±3.06 ab |
| 7 | 4 | 27 | 0.9 | 2 | 4.00±2.00 g |
| 8 | 4 | 28 | 0.7 | 3 | 21.33±6.43 efg |
| 9 | 5 | 25 | 0.7 | 5 | 68.33±12.42 a |
| 10 | 5 | 26 | 0.9 | 4 | 51.06±7.65 abc |
| 11 | 5 | 27 | 0.3 | 3 | 30.56±16.17 def |
| 12 | 5 | 28 | 0.5 | 2 | 4.00±5.29 g |
| 13 | 6 | 25 | 0.9 | 3 | 40.95±11.61 bcd |
| 14 | 6 | 26 | 0.7 | 2 | 8.52±1.70 g |
| 15 | 6 | 27 | 0.5 | 5 | 20.36±9.30 efg |
| 16 | 6 | 28 | 0.3 | 4 | 12.67±10.26 fg |
| K1 | 30.91 | 39.25 | 27.64 | 6.13 | |
| K2 | 31.09 | 37.49 | 23.78 | 30.97 | |
| K3 | 38.49 | 23.30 | 34.12 | 35.43 | |
| K4 | 20.62 | 21.07 | 35.58 | 48.58 | |
| R | 17.87 | 18.18 | 11.80 | 42.45 | |
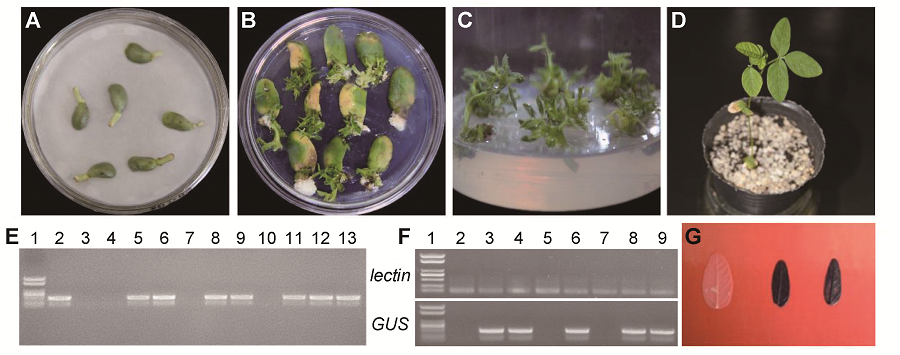
Figure 3 Transformation system of Glycine max cv. ‘Dongnong 50’ using optimized cotyledonary-node method(A) Co-cultivation after transformation; (B) Shoot induction; (C) Shoot elongation; (D) Resistant bud transplant; (E) PCR confirmation of GUS gene (1: Marker; 2: pCAMBIA3301-35S-GUS; 3: Control soybean; 4-13: Resistant plant); (F) RT-PCR confirmation of GUS gene (1: Marker; 2: Control soybean; 3-9: Resistant plant); (G) Histochemical staining of leaves from control (left) and resistant plant (middle and right)
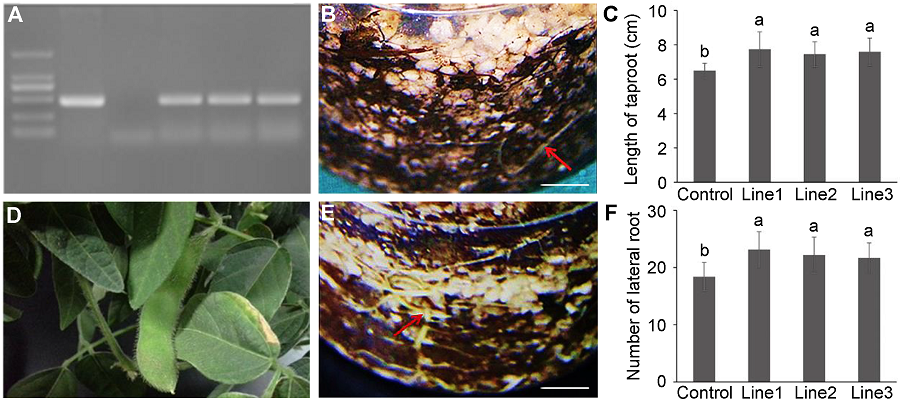
Figure 4 Confirmation and root system analyses of MtDREB2A transgenic soybean Dongnong 50(A) PCR result of MtDREB2A transgenic soybean (1: Marker; 2: pCAMBIA3301-rd29A-MtDREB2A; 3: Control soybean; 4-6: MtDREB2A transgenic soybean); (B) The root of control soybean (Bar=1 cm); (C) The length of taproot of control and transgenic soybeans; (D) The pod of MtDREB2A transgenic soybean in greenhouse; (E) The root of MtDREB2A transgenic soybean (Bar=1 cm); (F) The number of lateral root of control and transgenic soybeans. Different lowercase letters in Figures (C) and (F) indicate significant differences at 0.05 level according to Duncan’s test.
| Treatment | Factor | Adventitious bud induction rate (%) | Histochemical staining rate of adventitious bud (%) | ||||
|---|---|---|---|---|---|---|---|
| Recovery time (d) (A) | PPT concentration (mg·L-1) (B) | Cef concentration (mg·L-1) (C) | Cb concentration (mg·L-1) (D) | ||||
| 1 | 0 | 2 | 100 | 100 | 1.11±1.92 f | 0.00±0.00 | |
| 2 | 0 | 3 | 200 | 200 | 1.11±1.92 f | 1.11±1.92 | |
| 3 | 0 | 4 | 300 | 300 | 3.33±5.77 f | 2.22±1.92 | |
| 4 | 0 | 5 | 400 | 400 | 1.11±1.92 f | 1.11±1.92 | |
| 5 | 5 | 2 | 200 | 300 | 22.22±9.62 cd | 2.22±1.92 | |
| 6 | 5 | 3 | 100 | 400 | 7.78±3.85 ef | 3.33±0.00 | |
| 7 | 5 | 4 | 400 | 100 | 4.45±3.85 f | 1.11±1.92 | |
| 8 | 5 | 5 | 300 | 200 | 2.22±3.85 f | 1.11±1.92 | |
| 9 | 10 | 2 | 300 | 400 | 31.11±5.09 bcd | 0.00±0.00 | |
| 10 | 10 | 3 | 400 | 300 | 18.89±3.85 de | 0.00±0.00 | |
| 11 | 10 | 4 | 100 | 200 | 27.78±10.18 cd | 0.00±0.00 | |
| 12 | 10 | 5 | 200 | 100 | 21.11±8.39 cd | 1.11±1.92 | |
| 13 | 15 | 2 | 400 | 200 | 55.56±5.09 a | 0.00±0.00 | |
| 14 | 15 | 3 | 300 | 100 | 42.22±12.62 b | 0.00±0.00 | |
| 15 | 15 | 4 | 200 | 400 | 33.33±13.33 bc | 0.00±0.00 | |
| 16 | 15 | 5 | 100 | 300 | 23.33±8.82 cd | 0.00±0.00 | |
| Adventitious bud induction rate | K1 | 1.67 | 27.50 | 15.00 | 17.22 | ||
| K2 | 9.17 | 17.50 | 19.44 | 21.67 | |||
| K3 | 24.72 | 17.22 | 19.72 | 16.94 | |||
| K4 | 38.61 | 11.94 | 20.00 | 18.33 | |||
| R | 36.94 | 15.56 | 5.00 | 4.72 | |||
| Histochemical staining rate | K1 | 1.11 | 0.56 | 0.83 | 0.56 | ||
| K2 | 1.94 | 1.11 | 1.11 | 0.56 | |||
| K3 | 0.28 | 0.83 | 0.83 | 1.11 | |||
| K4 | 0.00 | 0.83 | 0.56 | 1.11 | |||
| R | 1.94 | 0.56 | 0.56 | 0.56 | |||
Table 6 The adventitious bud induction rate and histochemical staining rate of orthogonal experiment
| Treatment | Factor | Adventitious bud induction rate (%) | Histochemical staining rate of adventitious bud (%) | ||||
|---|---|---|---|---|---|---|---|
| Recovery time (d) (A) | PPT concentration (mg·L-1) (B) | Cef concentration (mg·L-1) (C) | Cb concentration (mg·L-1) (D) | ||||
| 1 | 0 | 2 | 100 | 100 | 1.11±1.92 f | 0.00±0.00 | |
| 2 | 0 | 3 | 200 | 200 | 1.11±1.92 f | 1.11±1.92 | |
| 3 | 0 | 4 | 300 | 300 | 3.33±5.77 f | 2.22±1.92 | |
| 4 | 0 | 5 | 400 | 400 | 1.11±1.92 f | 1.11±1.92 | |
| 5 | 5 | 2 | 200 | 300 | 22.22±9.62 cd | 2.22±1.92 | |
| 6 | 5 | 3 | 100 | 400 | 7.78±3.85 ef | 3.33±0.00 | |
| 7 | 5 | 4 | 400 | 100 | 4.45±3.85 f | 1.11±1.92 | |
| 8 | 5 | 5 | 300 | 200 | 2.22±3.85 f | 1.11±1.92 | |
| 9 | 10 | 2 | 300 | 400 | 31.11±5.09 bcd | 0.00±0.00 | |
| 10 | 10 | 3 | 400 | 300 | 18.89±3.85 de | 0.00±0.00 | |
| 11 | 10 | 4 | 100 | 200 | 27.78±10.18 cd | 0.00±0.00 | |
| 12 | 10 | 5 | 200 | 100 | 21.11±8.39 cd | 1.11±1.92 | |
| 13 | 15 | 2 | 400 | 200 | 55.56±5.09 a | 0.00±0.00 | |
| 14 | 15 | 3 | 300 | 100 | 42.22±12.62 b | 0.00±0.00 | |
| 15 | 15 | 4 | 200 | 400 | 33.33±13.33 bc | 0.00±0.00 | |
| 16 | 15 | 5 | 100 | 300 | 23.33±8.82 cd | 0.00±0.00 | |
| Adventitious bud induction rate | K1 | 1.67 | 27.50 | 15.00 | 17.22 | ||
| K2 | 9.17 | 17.50 | 19.44 | 21.67 | |||
| K3 | 24.72 | 17.22 | 19.72 | 16.94 | |||
| K4 | 38.61 | 11.94 | 20.00 | 18.33 | |||
| R | 36.94 | 15.56 | 5.00 | 4.72 | |||
| Histochemical staining rate | K1 | 1.11 | 0.56 | 0.83 | 0.56 | ||
| K2 | 1.94 | 1.11 | 1.11 | 0.56 | |||
| K3 | 0.28 | 0.83 | 0.83 | 1.11 | |||
| K4 | 0.00 | 0.83 | 0.56 | 1.11 | |||
| R | 1.94 | 0.56 | 0.56 | 0.56 | |||
| [1] | 薄路花, 曹越平 (2015). 不同大豆品种对农杆菌EHA105和GV3101敏感性及共培养条件的优化. 上海交通大学学报(农业科学版) 33, 26-31. |
| [2] | 董蕾, 任广明, 陈宝, 金羽, 曲娟娟 (2011). 转DREB基因大豆东农50对土壤氮素转化菌数量及生化强度的影响. 作物杂志 (5), 22-26. |
| [3] | 杜升伟, 刘业丽, 姚丙晨, 白晨, 苗兴芬, 刘春燕, 陈庆山, 胡国华 (2010). 大豆转化体系的优化和Dof4基因转入大豆的研究. 大豆科学 29, 398-402. |
| [4] | 杜艳丽, 谢甫绨 (2015). 转基因技术在大豆性状改良上的应用. 大豆科学 34, 320-328. |
| [5] | 段莹莹, 赵琳, 陈李淼, 李文滨 (2010). 农杆菌介导的大豆子叶节和下胚轴转化方法的比较及优化. 大豆科学 29, 590-593. |
| [6] | 韩献忠, 张治国, 刘骅, 赵立红 (1990). 条叶龙胆离体根培养条件的初步研究. 植物学通报 7(3), 49-51. |
| [7] | 侯文胜, 林抗雪, 陈普, 贾志伟, 周扬, 于洋, 刘雁华 (2014). 大豆规模化转基因技术体系的构建及其应用. 中国农业科学 47, 4198-4210. |
| [8] | 姜琼, 王幼宁, 王利祥, 孙政玺, 李霞 (2015). 盐胁迫下大豆根组织定量PCR分析中内参基因的选择. 植物学报 50, 754-764. |
| [9] | 林荣双, 梁丽琨, 肖显华, 王顺珍 (2003). 花生幼叶为外植体的植株再生系统的建立. 植物学通报 20, 307-312. |
| [10] | 林树柱, 曹越平, 卫志明, 马晓平, 陈鲁勇 (2005). 6-BA诱导大豆子叶节和茎尖出芽的研究. 上海交通大学学报(农业科学版) 23, 138-142. |
| [11] | 刘瑞江, 张业旺, 闻崇炜, 汤建 (2010). 正交试验设计和分析方法研究. 实验技术与管理 27(9), 52-55. |
| [12] | 刘银, 史秀岚, 王静磊, 刘琪迩, 王幼平 (2013). 大豆子叶节再生体系的建立. 扬州大学学报(农业与生命科学版) 34, 68-72. |
| [13] | 刘营, 张明辉, 霍楠, 仇有文, 敖金霞, 高学军 (2012). 转基因大豆OsDREB3品系特异性定性PCR检测方法的建立. 中国农业大学学报 17(4), 34-39. |
| [14] | 马晓红, 姚陆铭, 武天龙 (2008). 大豆整个子叶节外植体再生体系的建立及与子叶节、胚尖再生体系的比较. 大豆科学 27, 373-378. |
| [15] | 马艳, 肖娅萍, 王彩玲, 王哲之 (2004). 苦皮藤试管苗生根培养研究. 植物学通报 21, 332-336. |
| [16] | 邱波, 王志坤, 孟凡立, 李文滨 (2011). 不同大豆基因型再生性及对农杆菌敏感性的研究. 大豆科学 30, 752-756. |
| [17] | 桑庆亮, 赖钟雄, 林玉玲, 陈裕坤 (2014). 荔枝基因枪转化及其GUS瞬时表达研究. 热带作物学报 35, 2223-2229. |
| [18] | 王玲, 郭长奎, 任丁, 马红 (2017). 水稻非生物胁迫响应基因OsMIP1的表达与进化分析. 植物学报 52, 43-53. |
| [19] | 王志坤, Sebastian A, 常健敏, 李丹丹, 邱波, 张大勇, 李文滨 (2014). 转GmDof11基因高油转基因大豆的鉴定及主要农艺性状调查. 作物杂志 (2), 39-42. |
| [20] | 武小霞, 李静, 王志坤, 刘珊珊, 李海燕, 马永, 李文滨 (2010). 乙酰丁香酮浓度和共培养pH对大豆再生频率的影响. 东北农业大学学报 41(5), 1-4. |
| [21] | 修宇 (2016). FpDREB2A基因调控刺槐直根生长抗旱机制及选育改良抗旱优质材料基础研究. 博士论文. 北京: 北京林业大学. pp. 80-81. |
| [22] | 杨莹 (2013). 大豆中黄13农杆菌介导转化体系优化. 硕士论文. 北京: 北京林业大学. pp. 25-26. |
| [23] | 姚丙晨, 闫双勇, 苏京平, 马忠友, 王晓静, 孙玥, 刘学军 (2015). 大豆转基因研究进展. 大豆科技 (5), 18-26. |
| [24] | 袁鹰, 刘德璞, 郑培和, 温刚, 王玉民, 徐文静 (2004). 用基因枪将GUS基因导入玉米自交系的瞬时表达. 玉米科学 12, 41-43. |
| [25] | 赵团结, 盖钧镒 (2004). 栽培大豆起源与演化研究进展. 中国农业科学 37, 954-962. |
| [26] | Chen JR, Lü JJ, Liu R, Xiong XY, Wang TX, Chen SY, Guo LB, Wang HF (2010). DREB1C from Medicago truncatula enhances freezing tolerance in transgenic M. truncatula and China Rose(Rosa chinensis Jacq.). Plant Growth Regul 60, 199-211. |
| [27] | Chen JR, Lü JJ, Wang TX, Chen SY, Wang HF (2009). Activation of a DRE-binding transcription factor from Medicago truncatula by deleting a Ser/Thr-rich region. In Vitro Cell Dev Biol Plant 45, 1-11. |
| [28] | Donaldson PA, Simmonds DH (2000). Susceptibility to Agrobacterium tumefaciens and cotyledonary node trans- formation in short-season soybean. Plant Cell Rep 19, 478-484. |
| [29] | Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33, 751-763. |
| [30] | Gao MJ, Allard G, Byass L, Flanagan AM, Singh J (2002). Regulation and characterization of four CBF transcription factors from Brassica napus. Plant Mol Biol 49, 459-471. |
| [31] | Hao YJ, Wei W, Song QX, Chen HW, Zhang YQ, Wang F, Zou HF, Lei G, Tian AG, Zhang WK, Ma B, Zhang JS, Chen SY (2011). Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants.Plant J 68, 302-313. |
| [32] | Hinchee MAW, Connor-Ward DV, Newell CA, McDonnell RE, Sato SJ, Gasser CS, Fischhoff DA, Re DB, Fraley RT, Horsch RB (1988). Production of transgenic soybean plants using Agrobacterium-mediated DNA transfer. Nat Biotechnol 6, 915-922. |
| [33] | Hong HP, Zhang HY, Olhoft P, Hill S, Wiley H, Toren E, Hillebrand H, Jones T, Cheng M (2007). Organogenic callus as the target for plant regeneration and transformation viaAgrobacterium in soybean(Glycine max 43, 558-568. |
| [34] | Ko TS, Korban SS (2004). Enhancing the frequency of somatic embryogenesis followingAgrobacterium-media- ted transformation of immature cotyledons of soybean(Glycine max 40, 552-558. |
| [35] | Liu HK, Yang C, Wei ZM (2004). Efficient Agrobacterium tumefaciens-mediated transformation of soybeans using an embryonic tip regeneration system. Planta 219, 1042-1049. |
| [36] | Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis.Plant Cell 10, 1391-1406. |
| [37] | Olhoft P, Somers D (2001). L-cysteine increases Agrobacterium-mediated T-DNA delivery into soybean cotyledo- nary-node cells. Plant Cell Rep 20, 706-711. |
| [38] | Olhoft PM, Flagel LE, Donovan CM, Somers DA (2003). Efficient soybean transformation using hygromycin B selection in the cotyledonary-node method.Planta 216, 723-735. |
| [39] | Qin F, Kakimoto M, Sakuma Y, Maruyama K, Osakabe Y, Tran LSP, Shinozaki K, Yamaguchi-Shinozaki K (2007). Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J 50, 54-69. |
| [40] | Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression.Plant Cell 18, 1292-1309. |
| [41] | Seo JS, Sohn HB, Noh K, Jung C, An JH, Donovan CM, Somers DA, Kim DI, Jeong SC, Kim CG, Kim HM, Lee SH, Choi YD, Moon TW, Kim CH, Cheong JJ (2012). Expression of the Arabidopsis AtMYB44 gene confers drought/salt-stress tolerance in transgenic soybean. Mol Breed 29, 601-608. |
| [42] | Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003). Regulatory network of gene expression in the drought and cold stress responses.Curr Opin Plant Biol 6, 410-417. |
| [43] | Tran LSP, Quach TN, Guttikonda SK, Aldrich DL, Kumar R, Neelakandan A, Valliyodan B, Nguyen HT (2009). Molecular characterization of stress-inducible GmNAC genes in soybean. Mol Genet Genomics 281, 647-664. |
| [44] | Wang GL, Xu YN (2008). Hypocotyl-based Agrobacterium-mediated transformation of soybean(Glycine max) and application for RNA interference. Plant Cell Rep 27, 1177-1184. |
| [45] | Xiu Y, Iqbal A, Zhu C, Wu GD, Chang YP, Li N, Cao Y, Zhang WB, Zeng HM, Chen SY, Wang HF (2016). Improvement and transcriptome analysis of root architecture by overexpression of Fraxinus pennsylvanica DREB2A transcription factor in Robinia pseudoacacia L. ‘Idaho’. Plant Biotechnol J 14, 1456-1469. |
| [1] | Jiaxin Chen, Hao Mei, Caixiang Huang, Zongyuan Liang, Yitong Quan, Dongpeng Li, Buweimaieryemu•Saimaiti, Xinxin Li, Hong Liao. Establishment and Optimization of Chimeric Soybean Plant with Transgenic Hairy Roots [J]. Chinese Bulletin of Botany, 2024, 59(1): 0-0. |
| [2] | Yunhui Wang, Yifan Wang, Jiayu Lin, Jinhong Li, Shien Yao, Xiangchi Feng, Zhenlin Cao, Jun Wang, Meina Li. Plant Kinesin: from Microtubule Arrays to Physiological Regulation [J]. Chinese Bulletin of Botany, 2022, 57(3): 358-374. |
| [3] | Mengke Du, Wenting Lian, Xiao Zhang, Xinxin Li. Effects of Nitrogen Application on Nitrogen Fixation Capacity and GmLbs Expression in Soybean [J]. Chinese Bulletin of Botany, 2021, 56(4): 391-403. |
| [4] | Yan Wang, Bowei Jia, Mingzhe Sun, Xiaoli Sun. Advances in Molecular Mechanisms of Stress Tolerance in Wild Soybean [J]. Chinese Bulletin of Botany, 2021, 56(1): 104-115. |
| [5] | Zhengjun Xia, Yuzhuo Li, Jinlong Zhu, Hongyan Wu, Kun Xu, Hong Zhai. A Rapid, Non-destructive and Continuous Sampling Technique and DNA Extraction for Soybean Seed [J]. Chinese Bulletin of Botany, 2021, 56(1): 56-61. |
| [6] | Guangtao Zhu,Sanwen Huang. A 360-degree Scanning of Population Genetic Variations—a Pan-genome Study of Soybean [J]. Chinese Bulletin of Botany, 2020, 55(4): 403-406. |
| [7] | Feng Feng,Yong Zhan,Zhixi Tian. The Feasibility and Recommendation for Improving Soybean Production in Xinjiang [J]. Chinese Bulletin of Botany, 2020, 55(2): 199-204. |
| [8] | Kang Tang,Ruolin Yang. Origin and Evolution of Soybean Protein-coding Genes [J]. Chinese Bulletin of Botany, 2019, 54(3): 316-327. |
| [9] | Ai Wenqin, Jiang Hanyuan, Li Xinxin, Liao Hong. An Efficient Nutrient Solution System to Study Symbiotic Nitrogen Fixation in Soybean [J]. Chinese Bulletin of Botany, 2018, 53(4): 519-527. |
| [10] | Yan Li, Junyi Gai. The Genetic Basis of Soybean Extended to Tropical Regions [J]. Chinese Bulletin of Botany, 2017, 52(4): 389-393. |
| [11] | Zhengjun Xia. Research Progress in Whole-genome Analysis and Cloning of Genes Underlying Important Agronomic Traits in Soybean [J]. Chinese Bulletin of Botany, 2017, 52(2): 148-158. |
| [12] | Siyu Chen, Peng Liu, Mo Zhu, Dongdong Xia, Liang Li, Kezhang Xu, Zhanyu Chen, Zhian Zhang. Seed Vigor and Antioxidant Enzyme Activities During Germination in Different Canopies of Soybean [J]. Chinese Bulletin of Botany, 2016, 51(1): 24-30. |
| [13] | Wen Cheng, Zhengjun Xia, Xianzhong Feng, Suxin Yang. A Rapid and Nondestructive Method for Soybean DNA Extraction and Its Application [J]. Chinese Bulletin of Botany, 2016, 51(1): 68-73. |
| [14] | Qiong Jiang, Youning Wang, Lixiang Wang, Zhengxi Sun, Xia Li. Validation of Reference Genes for Quantitative RT-PCR Analysis in Soybean Root Tissue under Salt Stress [J]. Chinese Bulletin of Botany, 2015, 50(6): 754-764. |
| [15] | Candong Li, Tai Guo, Zhixin Wang, Wei Zheng, Zhenyu Zhang, Meiling Guo, Zhongtang Liu. Research into Nitrogen Application and Utilization Rate in Soybean Leaves With 15N Tracing Technique [J]. Chinese Bulletin of Botany, 2015, 50(4): 490-494. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||