

Chinese Bulletin of Botany ›› 2020, Vol. 55 ›› Issue (1): 83-89.DOI: 10.11983/CBB19213
• TECHNIQUES AND METHODS • Previous Articles Next Articles
Received:2019-10-30
Accepted:2019-12-11
Online:2020-01-01
Published:2019-12-20
Contact:
Gaoping Qu
Gaoping Qu,Jingbo Jin. Detection of SUMOylation in Plants[J]. Chinese Bulletin of Botany, 2020, 55(1): 83-89.
| Proteins | IPTG concentration (mmol·L-1) | Induction time |
|---|---|---|
| His-SUMO E1 | 1 | 16-20 h (16°C) |
| His-SUMO E2 | 1 | 3-5 h (28°C) |
| His-SUMO1GG | 1 | 16-20 h (16°C) |
| His-SUMO1AA | 1 | 16-20 h (16°C) |
| GST-protein X-Myc | 1 | 3-5 h (28°C) |
Table 1 Expression of SUMO reaction related proteins
| Proteins | IPTG concentration (mmol·L-1) | Induction time |
|---|---|---|
| His-SUMO E1 | 1 | 16-20 h (16°C) |
| His-SUMO E2 | 1 | 3-5 h (28°C) |
| His-SUMO1GG | 1 | 16-20 h (16°C) |
| His-SUMO1AA | 1 | 16-20 h (16°C) |
| GST-protein X-Myc | 1 | 3-5 h (28°C) |
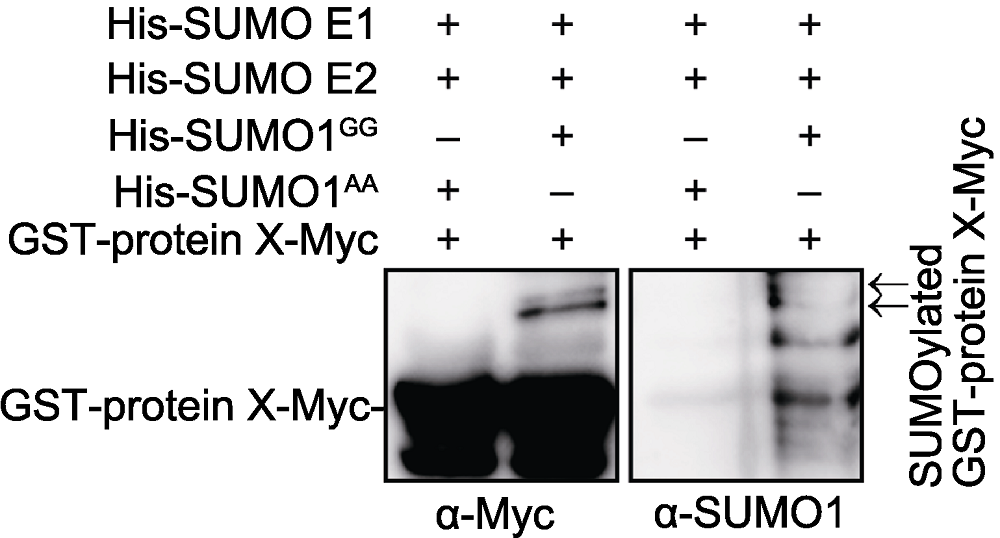
Figure 1 Protein X can be SUMOylated in vitro GST-protein X-Myc was incubated with His-SUMO E1, His- SUMO E2 and His-SUMO1GG (His-SUMO1AA was used as a negative control) at 30°C for 3 h. After reaction, GST-protein X-Myc was purified with Glutathione beads and detected with anti-Myc and anti-SUMO1 antibodies. Arrows represent SUMOylated GST-protein X-Myc.
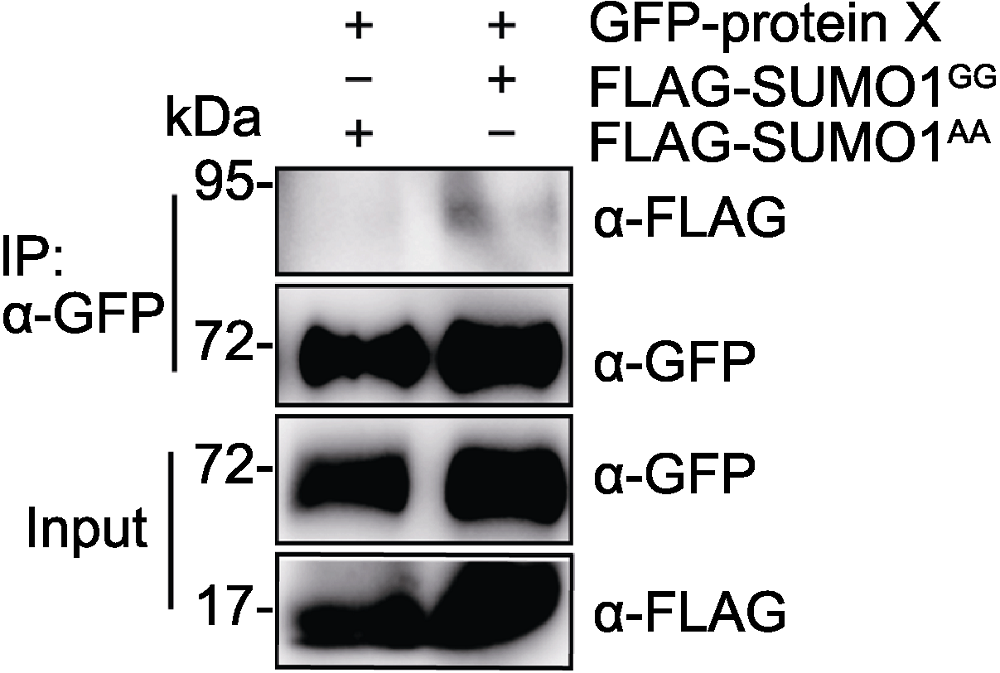
Figure 2 SUMOylation of GFP-protein X in Nicotiana benthamiana GFP-protein X was transiently co-expressed with FLAG-SUMO1GG or FLAG-SUMO1AA in Nicotiana benthamiana leaves. GFP-protein X was immunoprecipitated with anti-GFP antibody, and IP products were detected with anti-FLAG antibody.
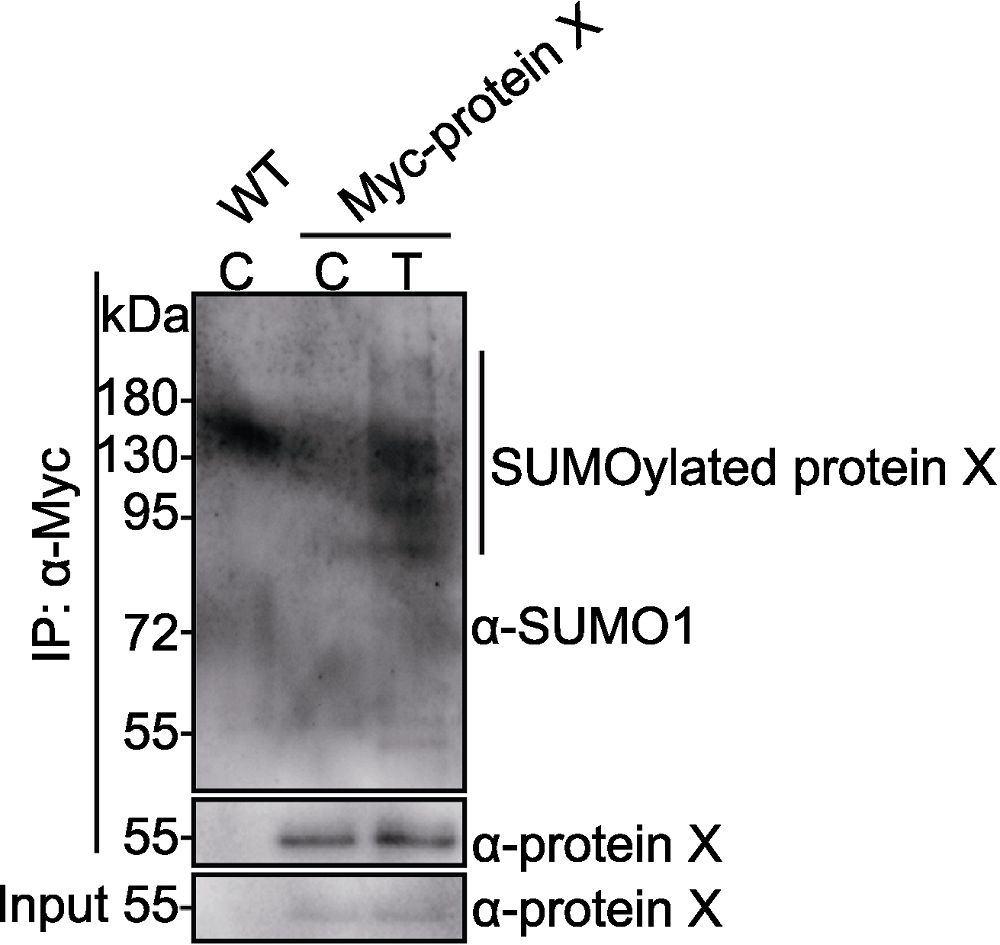
Figure 3 SUMOylation of Myc-protein X in transgenic plants Myc-protein X transgenic plants and wild-type (WT) were grown in control (C) or treatment (T) conditions for 3 days. Myc-protein X and SUMOylated Myc-protein X were detected with anti-protein X and anti-SUMO1 antibodies, respectively. Vertical line indicates SUMOylated Myc-protein X bands.
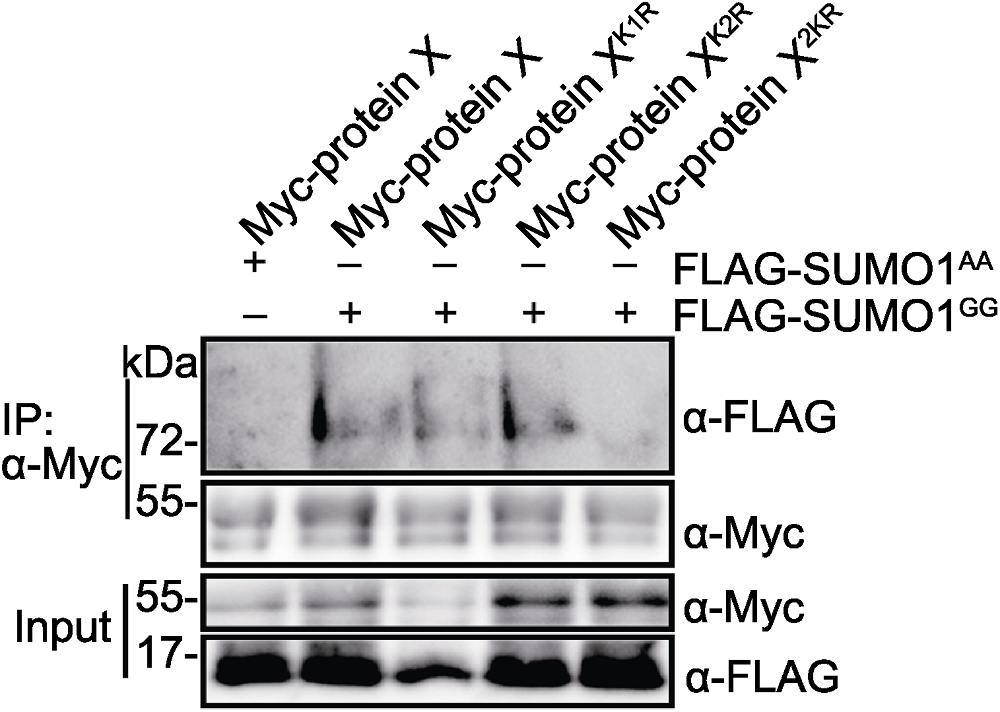
Figure 4 K1 and K2 are the primary SUMOylation sites of protein X Myc-protein X, Myc-protein XK1R, Myc-protein XK2R or Myc- protein X2KR was transiently co-expressed with FLAG-SUMO1GG or FLAG-SUMO1AA in Nicotiana benthamiana leaves, respectively.
| [1] | 徐庞连, 曾棉炜, 黄丽霞, 阳成伟 (2008). 植物SUMO化修饰及其生物学功能. 植物学通报 25, 608-615. |
| [2] | Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD (2002). Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108, 345-356. |
| [3] | Catala R, Ouyang J, Abreu IA, Hu YX, Seo H, Zhang XR, Chua NH (2007). The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19, 2952-2966. |
| [4] | Colby T, Matthäi A, Boeckelmann A, Stuible HP (2006). SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol 142, 318-332. |
| [5] | Conti L, Price G, O'Donnell E, Schwessinger B, Dominy P, Sadanandom A (2008). Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20, 2894-2908. |
| [6] | Elrouby N, Coupland G (2010). Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc Natl Acad Sci USA 107, 17415-17420. |
| [7] | Hermkes R, Fu YF, Nürrenberg K, Budhiraja R, Schmelzer E, Elrouby N, Dohmen RJ, Bachmair A, Coupland G (2011). Distinct roles for Arabidopsis SUMO protease ESD4 and its closest homolog ELS1. Planta 233, 63-73. |
| [8] | Ishida T, Yoshimura M, Miura K, Sugimoto K (2012). MMS21/HPY2 and SIZ1, two Arabidopsis SUMO E3 ligases, have distinct functions in development. PLoS One 7, e46897. |
| [9] | Kong XX, Luo X, Qu GP, Liu P, Jin JB (2017). Arabidopsis SUMO protease ASP1 positively regulates flowering time partially through regulating FLC stability. J Integr Plant Biol 59, 15-29. |
| [10] | Lin XL, Niu D, Hu ZL, Kim DH, Jin YH, Cai B, Liu P, Miura K, Yun DJ, Kim WY, Lin RC, Jin JB (2016). An Arabidopsis SUMO E3 ligase, SIZ1, negatively regulates photomorphogenesis by promoting COP1 activity. PLoS Genet 12, e1006016. |
| [11] | Liu LP, Jiang Y, Zhang XM, Wang X, Wang YB, Han YZ, Coupland G, Jin JB, Searle I, Fu YF, Chen FL (2017). Two SUMO proteases SUMO PROTEASE RELATED TO FERTILITY1 and 2 are required for fertility in Arabidopsis. Plant Physiol 175, 1703-1719. |
| [12] | Miller MJ, Barrett-Wilt GA, Hua ZH, Vierstra RD (2010). Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci USA 107, 16512-16517. |
| [13] | Murtas G, Reeves PH, Fu YF, Bancroft I, Dean C, Coupland G (2003). A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell 15, 2308-2319. |
| [14] | Rodriguez MS (2016). SUMO: Methods and Protocols . New York: Humana Press. |
| [15] | Rytz TC, Miller MJ, McLoughlin F, Augustine RC, Marshall RS, Juan YT, Charng YY, Scalf M, Smith LM, Vierstra RD (2018). SUMOylome profiling reveals a diverse array of nuclear targets modified by the SUMO ligase SIZ1 during heat stress. Plant Cell 30, 1077-1099. |
| [16] | Sadanandom A, Ádám É, Orosa B, Viczián A, Klose C, Zhang CJ, Josse EM, Kozma-Bognár L, Nagy F (2015). SUMOylation of phytochrome-B negatively regulates light- induced signaling in Arabidopsis thaliana. Proc Natl Acad Sci USA 112, 11108-11113. |
| [17] | Saleh A, Withers J, Mohan R, Marqués J, Gu YN, Yan SP, Zavaliev R, Nomoto M, Tada Y, Dong XN (2015). Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe 18, 169-182. |
| [18] | Saracco SA, Miller MJ, Kurepa J, Vierstra RD (2007). Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol 145, 119-134. |
| [19] | Tomanov K, Zeschmann A, Hermkes R, Eifler K, Ziba I, Grieco M, Novatchkova M, Hofmann K, Hesse H, Bachmair A (2014). Arabidopsis PIAL1 and 2 promote SUMO chain formation as E4-type SUMO ligases and are involved in stress responses and sulfur metabolism. Plant Cell 26, 4547-4560. |
| [20] | Yates G, Srivastava AK, Sadanandom A (2016). SUMO proteases: uncovering the roles of deSUMOylation in plants. J Exp Bot 67, 2541-2548. |
| [21] | Yoo SD, Cho YH, Sheen J (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2, 1565-1572. |
| [22] | Zhao Q, Xie YB, Zheng YY, Jiang S, Liu WZ, Mu WP, Liu ZX, Zhao Y, Xue Y, Ren J (2014). GPS-SUMO: a tool for the prediction of SUMOylation sites and SUMO-interaction motifs. Nucleic Acids Res 42, W325-W330. |
| [1] | Huang Junwen, Feng Qiyi, Zheng Kaiyong, Huang Junjie, Wang Linbo, Lai Jianbin Lai Ruiqiang, Yang Chengwei. An Effective in Vitro SUMOylation Detection System for Plant Proteins [J]. Chinese Bulletin of Botany, 2022, 57(4): 490-499. |
| [2] | Qiuxin Li, Wei Chi, Daili Ji. Research Progress of CURT1 on Regulating Thylakoid Membrane Curvature [J]. Chinese Bulletin of Botany, 2021, 56(4): 462-469. |
| [3] | Fei Du, Yuling Jiao. WUSCHEL-mediated Innate Immunity in Plant Stem Cells Provides a Novel Antiviral Strategy [J]. Chinese Bulletin of Botany, 2020, 55(5): 537-540. |
| [4] | Liang Wu, Yijun Qi. Small RNA, No Small Feat: Plants Deploy 22 nt siRNAs to Cope with Environmental Stress [J]. Chinese Bulletin of Botany, 2020, 55(3): 270-273. |
| [5] | Qingping Zhao,Shifan Ma,Ruixi Li,Tianyu Wang,Xiang Zhao. Advances of NPH3/RPT2-Like (NRL) Family Proteins in Phototropin-mediated Signaling in Arabidopsis thaliana [J]. Chinese Bulletin of Botany, 2020, 55(2): 240-253. |
| [6] | Hua Zhao,Guangda Shao,Wenxin Gao,Biao Gu. The Application of Double-barreled Particle Bombardment for Transient Gene Expression in Arabidopsis [J]. Chinese Bulletin of Botany, 2020, 55(2): 182-191. |
| [7] | Jing Zhang,Suiwen Hou. Role of Post-translational Modification of Proteins in ABA Signaling Transduction [J]. Chinese Bulletin of Botany, 2019, 54(3): 300-315. |
| [8] | Han Danlu, Lai Jianbin, Yang Chengwei. Research Advances in Functions of SUMO E3 Ligases in Plant Growth and Development [J]. Chinese Bulletin of Botany, 2018, 53(2): 175-184. |
| [9] | Panglian Xu;Mianwei Zeng;Lixia Huang;Chengwei Yang*. SUMOylation and Its Biological Function in Plants [J]. Chinese Bulletin of Botany, 2008, 25(05): 608-615. |
| [10] | Huixia Yang;Yiping Tong;Daowen Wang . Latest Advances in Understanding the Molecular Genetic Mechanism of Low Phosphate Responses in Arabidopsis thaliana [J]. Chinese Bulletin of Botany, 2007, 24(06): 726-734. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
