

Chinese Bulletin of Botany ›› 2018, Vol. 53 ›› Issue (1): 104-109.DOI: 10.11983/CBB16216
• TECHNIQUES AND METHODS • Previous Articles Next Articles
Yuan Cao, Yun Yang, Huaquan Xu, Yang Liu, Danyang Wan*( )
)
Received:2016-11-11
Accepted:2017-04-17
Online:2018-01-01
Published:2018-08-10
Contact:
Danyang Wan
Yuan Cao, Yun Yang, Huaquan Xu, Yang Liu, Danyang Wan. PCR Used to Find Plasmid Backbone Fragments in the Products of hiTAIL-PCR[J]. Chinese Bulletin of Botany, 2018, 53(1): 104-109.
| Primers | Sequence (5'-3') |
|---|---|
| Specific primers | |
| RB-S1 | GTTATCCGCTCACAATTCCACA |
| RB-S2 | TCGGGAAACCTGTCGTGCCA |
| RB-S3 | AGAGGCGGTTTGCGTATTGGG |
| RB-S4 | AAGTCGCTGTATGTGTTTGTTTGAGA |
| LB-A1 | GGCGGACCGCTATCAGGACAT |
| LB-A2 | TTGGCTACCCGTGATATTGCTG |
| LB-A3 | GACCGCTTCCTCGTGCTTTA |
| LB-A4 | GTTACACCACAATATATCCTGCCAAGAT |
| AC1 | ACGATGGACTCCAGAG |
| DRF1-S | ACAACAGAAACAACCAAAAATAATG |
| QRT-S | TGTGCAGGAGACATCATTCC |
| QRT-A | TTTCGCATTGCCAAAGAT |
| Random primers | |
| LAD1-1 | ACGATGGACTCCAGAVNVNNNGGAA |
| LAD1-2 | ACGATGGACTCCAGABNBNNGGTT |
| LAD1-3 | ACGATGGACTCCAGAVVNVNNNCCAA |
| LAD1-4 | ACGATGGACTCCAGABDNBNNNCGGT |
| LAD1-QA | ACGATGGACTCCAGAGWWWWHWWACCT |
| LAD1-HS | ACGATGGACTCCAGAGWWWWWWDYAGG |
| LAD1-A | ACGATGGACTCCAGAGVNVNNNGGCC |
| LAD1-B | ACGATGGACTCCAGAGBNBNNGGGG |
| LAD1-C | ACGATGGACTCCAGAGVVNVNNNCCGG |
| LAD1-D | ACGATGGACTCCAGAGBDNBNNNCCCC |
| LAD1-E | ACGATGGACTCCAGAGVNVNNNCAGA |
| LAD1-F | ACGATGGACTCCAGAGVNVNNNAGAT |
Table 1 Specific primers designed according to T-DNA sequence and random primers
| Primers | Sequence (5'-3') |
|---|---|
| Specific primers | |
| RB-S1 | GTTATCCGCTCACAATTCCACA |
| RB-S2 | TCGGGAAACCTGTCGTGCCA |
| RB-S3 | AGAGGCGGTTTGCGTATTGGG |
| RB-S4 | AAGTCGCTGTATGTGTTTGTTTGAGA |
| LB-A1 | GGCGGACCGCTATCAGGACAT |
| LB-A2 | TTGGCTACCCGTGATATTGCTG |
| LB-A3 | GACCGCTTCCTCGTGCTTTA |
| LB-A4 | GTTACACCACAATATATCCTGCCAAGAT |
| AC1 | ACGATGGACTCCAGAG |
| DRF1-S | ACAACAGAAACAACCAAAAATAATG |
| QRT-S | TGTGCAGGAGACATCATTCC |
| QRT-A | TTTCGCATTGCCAAAGAT |
| Random primers | |
| LAD1-1 | ACGATGGACTCCAGAVNVNNNGGAA |
| LAD1-2 | ACGATGGACTCCAGABNBNNGGTT |
| LAD1-3 | ACGATGGACTCCAGAVVNVNNNCCAA |
| LAD1-4 | ACGATGGACTCCAGABDNBNNNCGGT |
| LAD1-QA | ACGATGGACTCCAGAGWWWWHWWACCT |
| LAD1-HS | ACGATGGACTCCAGAGWWWWWWDYAGG |
| LAD1-A | ACGATGGACTCCAGAGVNVNNNGGCC |
| LAD1-B | ACGATGGACTCCAGAGBNBNNGGGG |
| LAD1-C | ACGATGGACTCCAGAGVVNVNNNCCGG |
| LAD1-D | ACGATGGACTCCAGAGBDNBNNNCCCC |
| LAD1-E | ACGATGGACTCCAGAGVNVNNNCAGA |
| LAD1-F | ACGATGGACTCCAGAGVNVNNNAGAT |
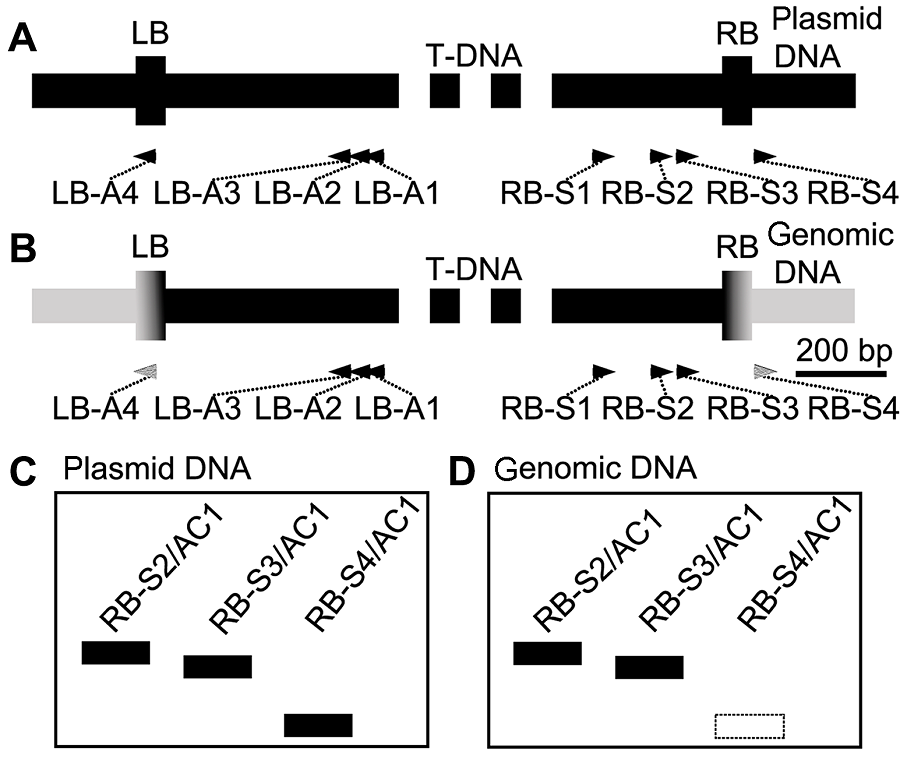
Figure 1 The principle of design(A) The regions of RB and LB of T-DNA plasmid; (B) The regions of RB and LB which integrate with chromosomal DNA; (C) When hiTAIL-PCR amplifies the backbone of plas- mid, the three PCR groups containing the RB-S2/AC1, RB-S3/AC1 and RB-S4/AC1 primer pairs, respectively, will all produce the positive bands; (D) When hiTAIL-PCR amplifies the genomic DNA, RB-S4/AC1 will not produce the positive bands (dashed frame)
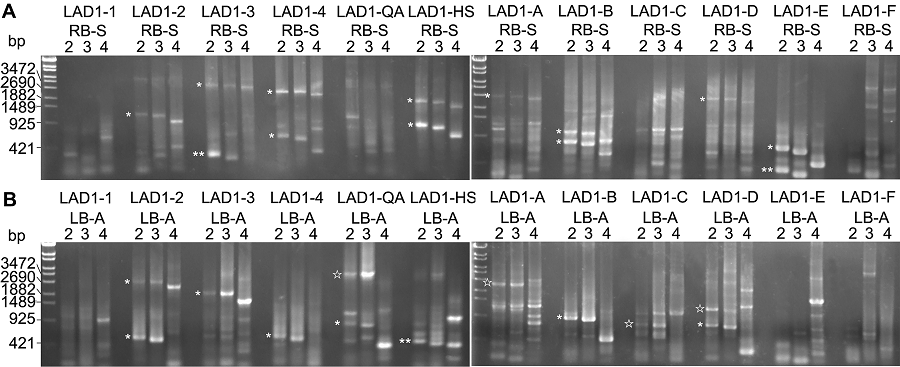
Figure 2 The cloning of flanking sequence at the T-DNA insertion site of Arabidopsis mutant drf1(A) The second round results amplified by PCR with RB-S serial primers; (B) The second round results amplified by PCR with LB-A serial primers. * display the non-specific amplification results; ☆ represent the potential specific amplification results; ** show the amplification results from the T-DNA region before RB or after LB.
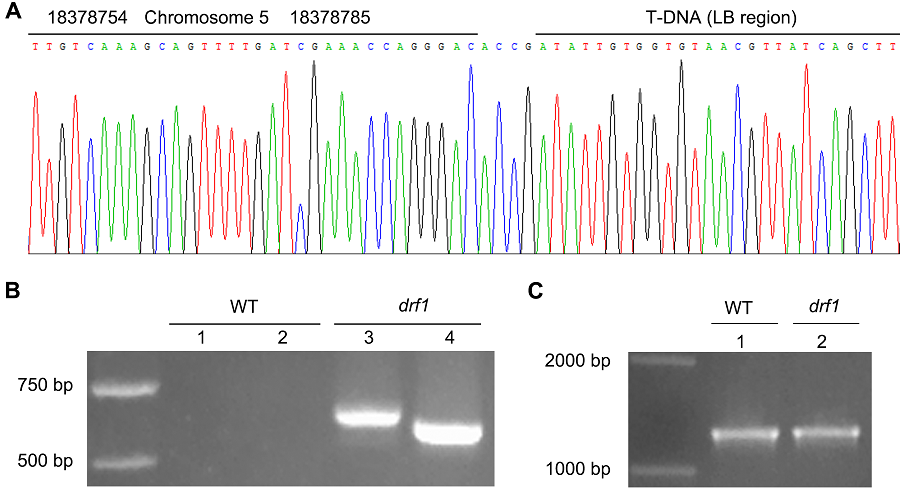
Figure 3 The PCR confirmation for Arabidopsis mutant drf1 flanking sequence(A) The partial sequencing result; (B) The results amplifying the genomic DNA from wild type (WT) and drf1 mutant with primer pairs of DRF1-S/LB-A2 (1,3) or DRF1-S/LB-A3 (2,4); (C) The PCR results with primer pairs of QRT-S/QRT-A to amplifying WT and drf1 mutant genomic DNA
| [1] | Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, Innan H, Cano L, Kamoun S, Terauchi R (2012). Genome sequencing reveals agronomically important loci in rice using MutMap.Nat Biotechnol 30, 174-178. |
| [2] | Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653-657. |
| [3] | Coulondre C, Miller JH (1977). Genetic studies of the lac repressor: III. Additional correlation of mutational sites with specific amino acid residues. J Mol Biol 117, 525-567. |
| [4] | Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42, 819-832. |
| [5] | Jander G (2006). Gene identification and cloning by molecular marker mapping.Methods Mol Biol 323, 115-126. |
| [6] | Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002). Arabidopsis map-based cloning in the post-genome era.Plant Physiol 129, 440-450. |
| [7] | Kleinboelting N, Huep G, Appelhagen I, Viehoever P, Li Y, Weisshaar B (2015). The structural features of thousands of T-DNA insertion sites are consistent with a double- strand break repair-based insertion mechanism.Mol Plant 8, 1651-1664. |
| [8] | Liu YG, Chen YL (2007). High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences.Biotechniques 43, 649-650. |
| [9] | Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8, 457-463. |
| [10] | Martineau B, Voelker TA, Sanders RA (1994). On defining T-DNA.Plant Cell 6, 1032-1033. |
| [11] | Mayerhofer R, Koncz-Kalman Z, Nawrath C, Bakkeren G, Crameri A, Angelis K, Redei GP, Schell J, Hohn B, Koncz C (1991). T-DNA integration: a mode of illegitimate recombination in plants.EMBO J 10, 697-704. |
| [12] | Mueller PR, Wold B (1989). In vivo footprinting of a muscle specific enhancer by ligation mediated PCR.Science 246, 780-786. |
| [13] | Nan GL, Walbot V (2009). Plasmid rescue: recovery of flanking genomic sequences from transgenic transposon insertion sites.Methods Mol Biol 526, 101-109. |
| [14] | Riley J, Butler R, Ogilvie D, Finniear R, Jenner D, Powell S, Anand R, Smith JC, Markham AF (1990). A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones.Nucleic Acids Res 18, 2887-2890. |
| [15] | Stachel SE, Timmerman B, Zambryski P (1987). Activation of Agrobacterium tumefaciens vir gene expression generates multiple single-stranded T-strand molecules from the pTiA6 T-region: requirement for 5' virD gene products. EMBO J 6, 857-863. |
| [16] | Triglia T, Peterson MG, Kemp DJ (1988). A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res 16, 8186. |
| [17] | Tzfira T, Li JX, Lacroix B, Citovsky V (2004). Agrobacterium T-DNA integration: molecules and models.Trends Ge- net 20, 375-383. |
| [18] | Wang HR, Fang J, Liang CZ, He MH, Li QY, Chu CC (2011). Computation-assisted SiteFinding-PCR for isolating flanking sequence tags in rice.Biotechniques 51, 421-423. |
| [19] | Zhou ZW, Ma HY, Qu LJ, Xie F, Ma QW, Ren ZR (2012). Establishment of an improved high-efficiency thermal asym- metric interlaced PCR for identification of genomic integration sites mediated by phiC31 integrase.World J Microbiol Biotechnol 28, 1295-1299. |
| [1] | Kairu Yang, Qiwei Jia, Jiayi Jin, Hanfei Ye, Sheng Wang, Qianyu Chen, Yian Guan, Chenyang Pan, Dedong Xin, Yuan Fang, Yuexing Wang, Yuchun Rao. Cloning and Functional Analysis of Rice Yellow Green Leaf Regulatory Gene YGL18 [J]. Chinese Bulletin of Botany, 2022, 57(3): 276-287. |
| [2] | Xia Wang, Wei Yan, Zhiqin Zhou, Zhenyi Chang, Minting Zheng, Xiaoyan Tang, Jianxin Wu. Identification and Mapping of a Rice Male Sterility Mutant ms102 [J]. Chinese Bulletin of Botany, 2022, 57(1): 42-55. |
| [3] | Jiangyuan Shang, Yan Chun, Xueyong Li. Map-based Cloning and Natural Variation Analysis of the PAL3 Gene Controlling Panicle Length in Rice [J]. Chinese Bulletin of Botany, 2021, 56(5): 520-532. |
| [4] | Jing Lu, Yingnan Chen, Tongming Yin. Research Progress on Sex Determination Genes of Woody Plants [J]. Chinese Bulletin of Botany, 2021, 56(1): 90-103. |
| [5] | Xifeng Chen,Yaping Liu,Bojun Ma. Design and Practice of a New Teaching Project of the Map-based Cloning Experiment in Genetics [J]. Chinese Bulletin of Botany, 2019, 54(6): 797-803. |
| [6] | Dewei Yang,Mo Wang,Libo Han,Dingzhong Tang,Shengping Li. Progress of Cloning and Breeding Application of Blast Resistance Genes in Rice and Avirulence Genes in Blast Fungi [J]. Chinese Bulletin of Botany, 2019, 54(2): 265-276. |
| [7] | Wang Yunqian, Su Yanhong, Yang Rui, Li Xin, Li Jing, Zeng Qianchun, Luo Qiong. Rice Blast Resistance of Wild Rice in Yunnan [J]. Chinese Bulletin of Botany, 2018, 53(4): 477-486. |
| [8] | Li Ma, Wancang Sun, Jinhai Yuan, Zigang Liu, Junyan Wu, Yan Fang, Yaozhao Xu, Yuanyuan Pu, Jing Bai, Xiaoyun Dong, Huili He. Expression Analysis of β-1,3-Glucanase Gene from Winter Brassica rapa Under Low Temperature Stress [J]. Chinese Bulletin of Botany, 2017, 52(5): 568-578. |
| [9] | Zhengjun Xia. Research Progress in Whole-genome Analysis and Cloning of Genes Underlying Important Agronomic Traits in Soybean [J]. Chinese Bulletin of Botany, 2017, 52(2): 148-158. |
| [10] | Wang Wenle, Feng Dan, Wu Jinxia, Zhang Zhiguo, Lu Tiegang. Gene Mapping of a Dwarf Gene WLD1 in Rice [J]. Chinese Bulletin of Botany, 2017, 52(1): 54-60. |
| [11] | Danlong Jing, Yan Xia, Shougong Zhang, Junhui Wang. Expression Analysis of B-class MADS-box Genes from Catalpa speciosa [J]. Chinese Bulletin of Botany, 2016, 51(2): 210-217. |
| [12] | Yongshu Liang, Junjie Zhou, Wenbin Nan, Dongdong Duan, Hanma Zhang. Progress in Rice Root System Research [J]. Chinese Bulletin of Botany, 2016, 51(1): 98-106. |
| [13] | Lingfei Li, Jianzong Peng, Xiaojing Wang. Preliminary Functional Analysis of Microtubule-associated Protein GMAP65-1 from Gerbera hybrida [J]. Chinese Bulletin of Botany, 2015, 50(1): 12-21. |
| [14] | Yuanbao Cai, Xiangyan Yang, Guangming Sun, Qiang Huang, Yeqiang Liu, Shaopeng Li, Zhili Zhang. Cloning of Flowering-related Gene AcMADS1 and Characterization of Expression in Tissues of Pineapple (Ananas comosus) [J]. Chinese Bulletin of Botany, 2014, 49(6): 692-703. |
| [15] | Yujing Wang, Minchun Li, Wang Wu, Hanying Wu, Yinong Xu. Cloning and Characterization of an AP2/EREBP Gene TmAP2-1 from Tetraena mongolica [J]. Chinese Bulletin of Botany, 2013, 48(1): 23-33. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||